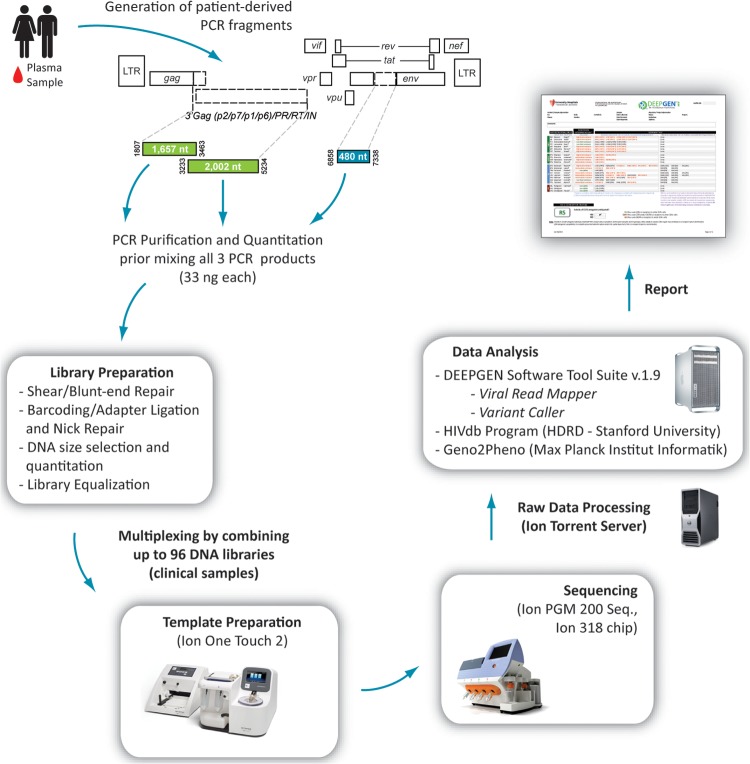
FIG 1.
Overview of the protocol for the novel HIV-1 genotyping and coreceptor tropism assay (DeepGen HIV). Three PCR products corresponding to the gag-p2/NCp7/p1/p6/pol-PR/RT/IN- (1,657 bp and 2,002 bp) and env-C2V3- (480 bp) coding regions of HIV-1 were used to construct a multiplexed library for shotgun sequencing on the Ion PGM. Signal processing and base calling were performed with the Torrent analysis suite version 3.4.2 and sequences analyzed using DEEPGEN Software Tool Suite. The HIVdb Program Genotypic Resistance Interpretation Algorithm from the Stanford University HIV Drug Resistance Database (see http://hivdb.stanford.edu) and Geno2Pheno (27) were used to infer the levels of susceptibility to PI, RTI, and INI and for HIV-1 coreceptor tropism determination, respectively.