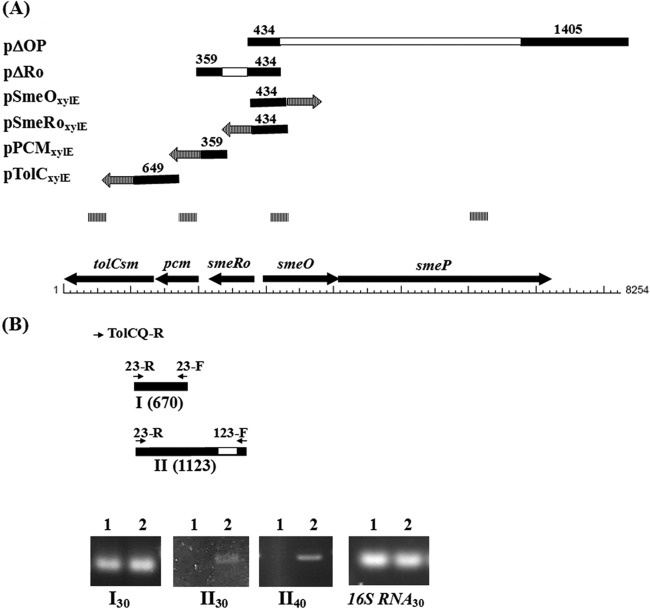
FIG 1.
Schematic organization of the smeOP and smeRo-pcm-tolCSm operons from S. maltophilia strain KJ. The smeOP operon contains genes for a membrane fusion protein (SmeO) and an RND transporter (SmeP). A TetR-type transcriptional regulator gene, smeRo, is located immediately upstream of the smeOP operon and is divergently transcribed. The smeRo, pcm, and tolCSm genes form an operon. Solid arrows represent open reading frames (ORFs) and the direction of transcription. (A) Structures of recombinant plasmids. The solid lines represent the PCR amplicons, and each empty bar represents the deleted region for each plasmid construct. The numbers above the solid bars represent the PCR amplicon sizes (in bp). The arrows with vertical lines indicate the xylE gene. The vertical-line bars indicate the products of qRT-PCR. (B) Presence of the smeRo-pcm-tolCSm operon. The solid bars labeled I and II indicate the products of RT-PCR generated by primers 23-F/23-R and 123-F/23-R, respectively. The numbers in parentheses represent the PCR amplicon sizes (in bp). The arrows indicate the positions of primers. The empty bar represents the deleted region in strain KJΔRo. The agarose gels show the RT-PCR products of strains KJ and KJΔRo. Lanes 1, strain KJ; lanes 2, strain KJΔRo. The RT-PCR products, labeled I and II, were generated by primers 23-F/23-R and 123-F/23-R, respectively. The subscript numbers indicate the numbers of PCR cycles.