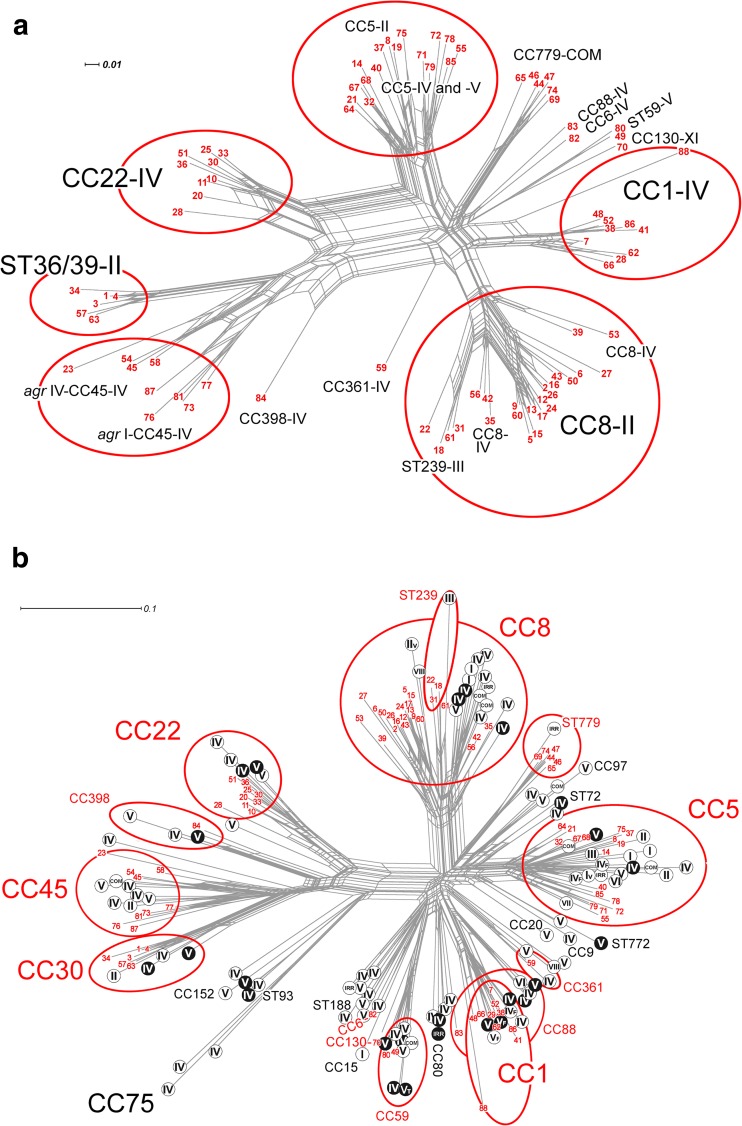
FIG 1.
Network trees generated using the SplitsTree, version 4.11.3, software program (1, 35) and the StaphyType DNA microarray profiles as described previously (1) to visualize the similarities and relationships between CCs and mobile genetic elements, including SCCmec, for the 88 sporadic MRSA isolates investigated and a global population of MRSA isolates. (a) Network tree showing the relationships between the 88 sporadic MRSA isolates investigated. (b) Network tree showing the relationships between the 88 sporadic MRSA isolates investigated in the present study and a previously described global collection of MRSA isolates (n = 3,139) (1). Each sporadic MRSA isolate investigated in the present study is indicated with a number in red (numbers 1 to 88); the details of isolates represented by each number are listed in Table 1. The major CCs identified among isolates in the current study and the previous global study are circled in red, and Roman numerals indicate SCCmec types. In panel b, CCs and STs that were exhibited by MRSA strains from this study are shown in red, and if a CC or ST was not exhibited by any of the 88 sporadic MRSA strains, it is shown in black. MRSA strains from the previously described global population (1) that lacked the Panton-Valentine leukocidin toxin genes lukF/S-PV (pvl) are indicated using black letters on a white background, and pvl-positive MRSA strains are indicated using white letters on a black background. The scale bars show how the length of a branch translates in sequence divergence. The unit is divergent nucleotides divided by the length of the sequence analyzed. Abbreviations: IRR, irregular SCCmec elements; COM, composite or multiple SCCmec elements.