Abstract
A multimodal acidic organelle targeting activity-based probe was developed to measure subcellular native enzymatic activity of cells by fluorescent microscopy and mass spectrometry. A cathepsin reactive warhead, conjugated to a weakly basic amine and a clickable alkyne, for subsequent appendage of a fluorophore or biotin reporter tag, accumulated in lysosomes as observed by Structured Illumination Microscopy (SIM) in J774 mouse macrophage cells. Analysis of in vivo labeled J774 by mass spectrometry showed that the probe was very selective for Cathepsins B and Z, two lysosomal cysteine proteases. Analysis of starvation-induced autophagy, a catabolic pathway involving lysosomes, showed a large increase in tagged protein number and an increase in cathepsin activity. Organelle targeting activity-based probes, enabled by fine-tuning of physicochemical features, holds great promise for characterizing enzyme activities in the myriad diseases implicated to subcellular locales, particularly the lysosome.
Keywords: proteomics, mass spectrometry, activity-based probe, lysosome, fluorescent microscopy
Activity-Based Protein Profiling (ABPP) utilizes targeted chemical probes to determine enzyme activities in vitro, in situ, and in vivo.[1] Employing probe derivatives containing an azide or alkyne moiety permits multiple applications of ABPP, such as fluorescent microscopy and mass spectrometry (MS), by direct attachment of reporting groups using copper-catalyzed azide-alkyne cycloadditions (CuAAC).[2] Small probe size increases cell permeability thereby allowing interrogation of the native proteome within intact cells. Using ABPP to irreversibly bind shared catalytic features of soluble enzyme families within a defined organelle population is an exceptional challenge for chemoproteomics.[3] Organelle proteomics is plagued by difficulties in distinguishing true resident proteins from co-purifying contaminant proteins, and cellular disruption for organelle isolation potentially interferes with native enzymatic activity.[4] Thus, we endeavored to analyze specific active enzyme populations in defined organelles within live cells using cell permeable organelle-directed activity-based probes (ABP).
We envisioned an acidotropic ABP that would accumulate in acidic organelles for interrogation of resident enzyme family activity. High molecular weight probes containing fluorophores and/or enrichment tags are known, but require cell penetrating peptides or receptor motifs, in addition to long labeling times, in order to enter live cells and accumulate.[5] Smaller ABPs can reach their enzyme target quickly, but any organelle accumulation is due to enzyme localization rather than defined probe properties.[2b, 6] Our envisioned ABP requires a moiety with specific physicochemical properties,[7] and an alkyne for CuAAC mediated conjugation of a fluorophore or enrichment tag.
For proof-of-concept, we designed a lysosome-targeting ABP. Lysosomes are ubiquitous in mammalian cells, important for cell homeostasis, and have roles in cancer, Alzheimer’s disease, and numerous orphan diseases.[8] Lysosomes are acidic organelles (pH < 5) that contain at least 60 soluble hydrolase enzymes. Lysosomal proteomics is increasingly important for the study of lysosome associated diseases, and the discovery of novel enzymes.[8] Lysosomes are targeted using weakly basic amines and lipophilic moieties, which accumulate in acidic environments due to passive diffusion of a neutral species across the membrane into an acidic milieu where the weak base is protonated thereby preventing diffusion back across the membrane.[9]
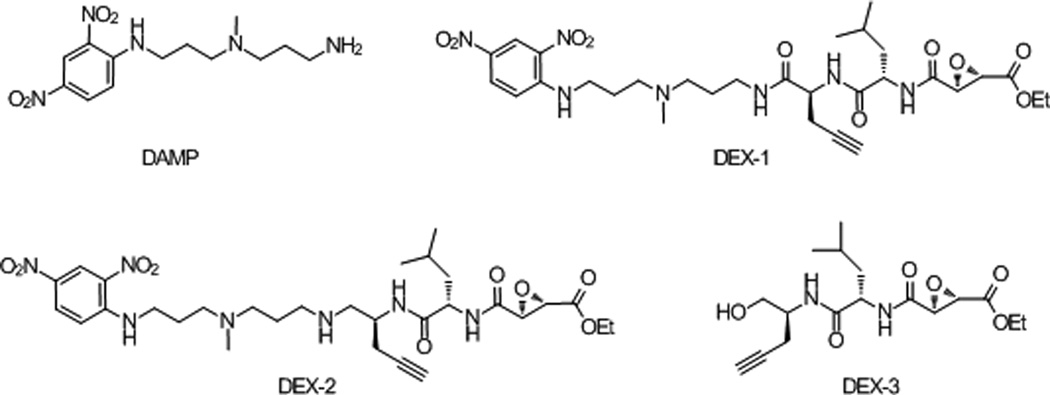
We targeted acidic organelles by incorporating readily available 3-(2,4-dinitroanilino)-3'-amino-N-methyldipropylamine (DAMP) into an ABP (Figure 1). DAMP is known to rapidly accumulate in acidic organelles due to its primary amine and pKa of ~10.[10] Furthermore, the DAMP primary amine provides a sufficient handle to append 1) an alkyne for CuAAC,[11] and 2) the broadly reactive papain cysteine protease inhibitor ethyl succinate epoxide, extensively used for labeling cathepsins.[2b, 5–6, 11] The synthesis of organelle-targeting DAMP epoxide probes (DEX-1 and DEX-2) is described in the supporting information (Figure 1, Figure S1 and S2). An ABP lacking DAMP (DEX-3) was synthesized as a weak acid negative control (Figure 1, Figure S2).[9] We suspected that disruption of an intramolecular hydrogen bond between the basic amines of DAMP by replacement with the amide of DEX-1 would prevent efficient accumulation in acidic organelles; therefore, we synthesized amine derivative DEX-2.
Figure 1.
DAMP derived, lysosome-targeting activity-based probes.
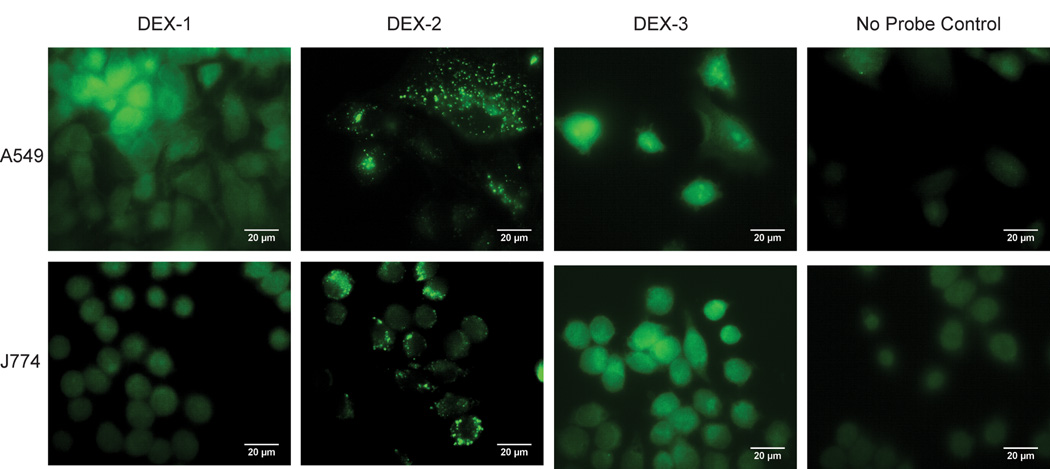
We applied our suite of organelle targeting probes to A549 – human epithelial cells, and J774 mouse macrophages (Figure 2). Imaging reveals DEX-2 accumulation in punctate vesicles throughout A549 and J774, while DEX-3 and DEX-1 diffusely label (Figure 2). The more basic DEX-2 accumulates in punctate vesicles quickly and permanently (Figure S3), with little cytosol labeling. All ABPs react with similar nucleophilic proteases in A549 global cell lysate (Figure S4A); additionally, comparison of in vivo J774 labeling shows that DEX-1 and DEX-2 have similar intracellular targets except for two intense bands of low molecular weight (MW, <30 kDa, Figure S4B). Thus, selective labeling of these low MW proteins and accumulation of DEX-2 is dependent on the basic amine.
Figure 2.
A549 and J774 cell lines were incubated with the DEX probe suite (10 µM and 2 µM, respectively, 1h), fixed, and conjugated to AlexaFluor 488 via CuAAC. DEX-2 shows punctate vesicle staining in both cell lines. DEX-3 and DEX-1 diffusely label throughout the cell. No probe control corresponds to DMSO treatment followed by CuAAC to the fluorophore.
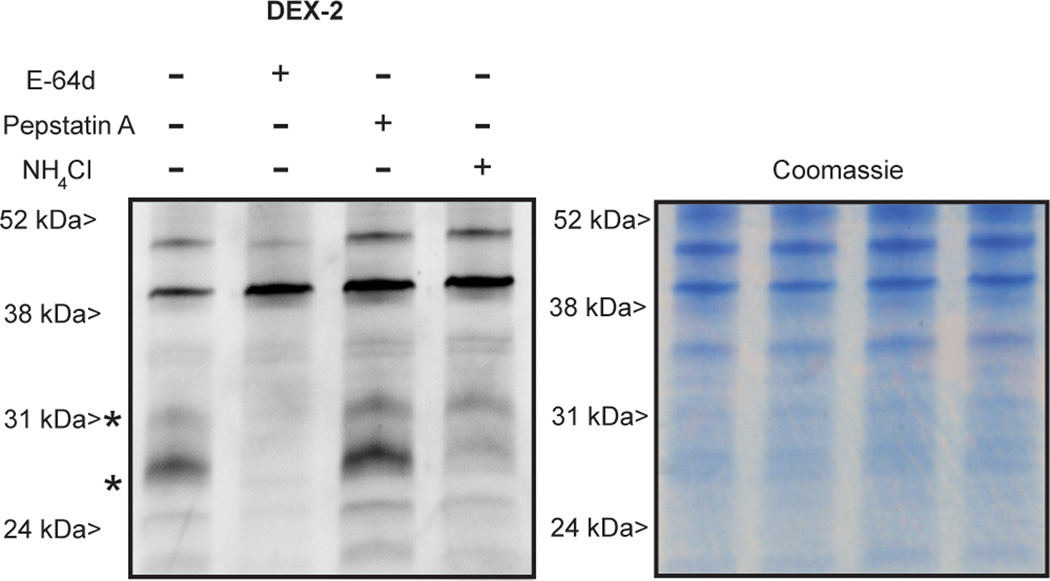
Accumulation of DEX-2 is dependent on the acidic environment of the target organelle and the activity of the target enzyme. Pretreatment of J774 with excess NH4Cl, known to neutralize acidic environments, reduced labeling intensity of punctate vesicles (Figure S5). Additionally pretreatment of cells with iodoacetamide (IAM) or ethyl (2S,3S)-oxirane dicarboxylate reduced labeling intensity (Figure S5). The fluorescent imaging experiments are consistent with gel based analysis; pretreatment of J774 cells with NH4Cl and a slight excess of the cathepsin inhibitor E-64d suppresses labeling of bands at 52 kDa, 31 kDa, and 27 kDa suggesting that these proteins are cathepsins and that labeling is dependent on the acidic environment (Figure 3). The aspartyl protease inhibitor pepstatin A did not suppress labeling. DEX-2 labels purified cathepsin B and J774 proteins in a dose-dependent manner (Figure S6). Notably, the intense band at ~40 kDa appears to be a result of CuAAC (Figure S7).
Figure 3.
Fluorescent imaging of J774 cells pre-treated with E-64d (58 µM), pepstatin A (58 µM), or NH4Cl (30 mM) and then labeled in vivo with DEX-2(37 µM). The Coomassie protein stain shows equivalent protein loading of gel lanes. Western blot analysis and LC-MS of J774 labeled in vivo with DEX-2 confirms labeling of CatB (28 kDa) and CatZ (27 kDa). Figure S7 shows a gel of in vivo DEX-2 labeling versus a no probe control.
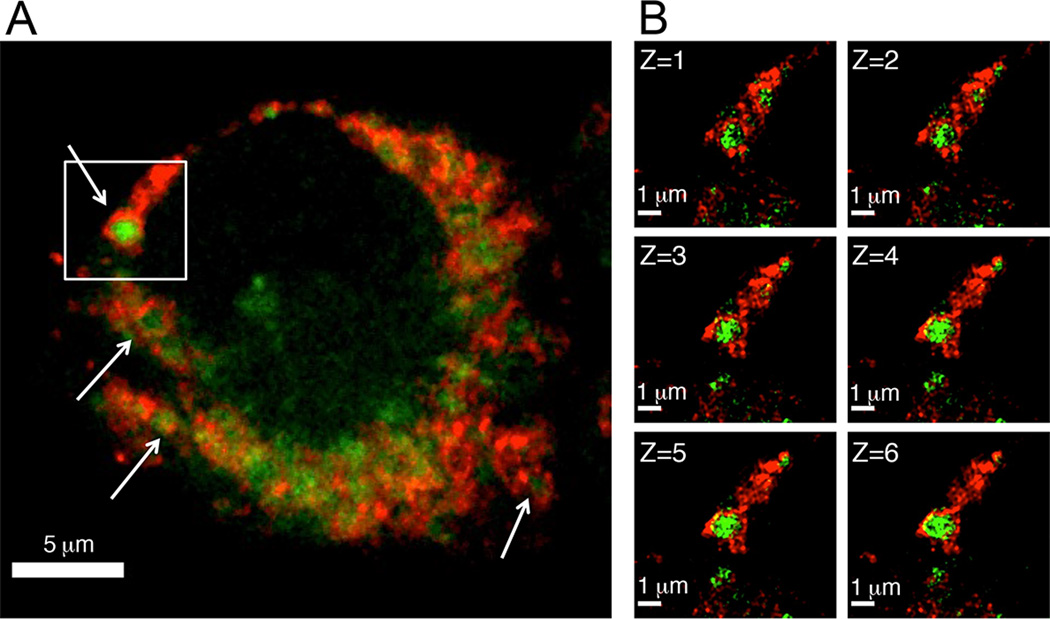
To identify the site of DEX-2 accumulation, J774 cells treated with DEX-2 were immunostained for LAMP1, a lysosome membrane marker (Figure 4A). Confocal laser scanning microscopy showed partial co-localization of DEX-2 with LAMP1 (Pearson’s correlation coefficient (PCC) = 0.595 ± 0.082, M1 = 0.919 ± 0.026, M2 = 0.958 ± 0.014)[12] (Figure 4A). In fact it appears that a LAMP1 membrane ring encases the fluorescent signal of DEX-2 (arrows, Figure 4A).[13] This invagination of probe signal by LAMP1 is clearly present at resolutions below the diffraction limits of light (<200 nm) using super-resolution Structured Illumination Microscopy (SIM, Figure 4B).[14] Z-stack imaging revealed that the phenomenon is conserved throughout the vesicle (Figure 4B; 3-D SI Videos S1 & S2), demonstrating that the probe is engulfed within the lysosome.
Figure 4.
Co-localization of J774 cells labeled with DEX-2 (green) and LAMP1 (red). J774 cells were treated with DEX-2 (2 µM) for 1 h. Cells were fixed and probe was conjugated to AlexaFluor 488. Lysosomes were stained by immunofluorescence with anti-LAMP1, goat anti-rabbit IgG-Alexafluor 647. A) Confocal laser scanning microscopy shows the DEX-2 signal encased by the LAMP1 antibody red signal, marked by arrows. B) The boxed region of the confocal image was analyzed further by enhanced resolution SIM. Shown are the vertical Z-plane images. Starting from the top of the compartment (Z=1) to the middle of the compartment (Z=6); each image is a 110 nm step (see also 3-D SI Videos S1 & S2).
We performed high resolution LC-MS/MS analysis of J774 cells treated with DEX-2 to characterize enzyme targets under two growth conditions. No probe DMSO control experiments were run in parallel to account for background labeling. We identified six proteins in healthy J774 cells (Table S1). Cathepsins B and Z were the most abundant proteins measured. J774 cells were also labeled under starvation-induced autophagy to determine if lysosome-targeting DEX-2 could report on enzyme activity changes during stress (Figure S8). Autophagy, the digestion and recycling of cellular components such as organelles and intracellular proteins,[15] is important for cell homeostasis and response to stress, and has been identified as having a role in cancer and Alzheimer’s disease.[16] Studies have shown that the expression and/or efficiency of hydrolytic enzymes can change during autophagy.[17] Furthermore, DAMP accumulates in autolysosomes, a fused lysosome autophagosome where degradation occurs.[18] Using the same data filtering criteria, DEX-2 labeled 44 proteins during autophagy (Table S1). Previously, 17 of the 44 proteins were observed in autophagosomes under nitrogen starvation-induced autophagy in human MCF7-eGFP-LC by stable isotope labeling by amino acids in cell culture (SILAC) and protein correlation profiling analysis.[19]
Autophagy constitutes degradation of cellular proteins and organelles in autolysosomes. Consistent with this, DEX-2 labeled proteins in autophagic cells come from a range of cellular locales including cytosol, mitochondria, actin cytoskeleton, and ER. Previously, a global SILAC proteomics analysis of amino acid starved MCF7-eGFP-LC3 cell lysate found that proteins associated with these locations decreased in abundance to different extents over a 36 h period.[20] Our observation of these proteins within acidic organelles after only 1 h of starvation showcases the sensitivity of our organelle-targeting ABP method. The loss of selectivity observed for cysteine proteases in autophagic cells by DEX-2 is likely due to nucleophilic residue exposure resulting from partial degradation of these proteins during autophagy.
We observed that enzyme activity increases during autophagy. Cathepsins B and Z had a moderate increase of 1.6-fold (p-value < 0.05). Previous studies show that short-term starvation and amino acid deprivation induce proteolysis.[13, 17b, 21] Furthermore, in mouse liver, cathepsin B activity initially decreases but then increases over 68 h of starvation.[21] These enzyme activity assays were performed on liver homogenate and may not accurately portray enzyme activity in a live native cellular environment. The small activity increase we measured for the cathepsins correlates with the very short starvation interval we used to induce autophagy for this study, and the transient nature of autolysosomes in autophagic flux. It also illustrates that Cathepsin B enzymatic activity is variable and contingent upon conditions.
In conclusion, we developed a multimodal organelle-targeting ABP that rapidly accumulates in punctate vesicles due to the physicochemical characteristics of a weakly basic amine. Using super-resolution SIM we provide evidence that these punctate vesicles are lysosomes. The probe was also used to measure enzyme activity changes upon induction of autophagy in live macrophage cells. Direction of an ABP to organelles by physicochemical properties should be applicable to other organelles. Additionally, switching the electrophilic warhead of a defined organelle targeting ABP would allow targeting of other resident enzyme families. We believe this approach can fuse organelle and activity-based proteomics, and that it can be widely applicable for analysis of biological systems.
Supplementary Material
Footnotes
This work was supported in part by the Laboratory Directed Research and Development Program at PNNL, a multiprogram national laboratory operated by Battelle for the U.S. DOE under Contract DE-AC05-76RL01830, and by the NIH NIGMS (8 P41 GM103493-11). SDW was supported by the PNNL Linus Pauling Distinguished Postdoctoral Fellowship. Work was performed in the Environmental Molecular Sciences Laboratory, a US DOE-BER national scientific user facility at PNNL. This work used instrumentation and capabilities developed under support from the NIH (8 P41 GM103493-11) and the DOE-BER.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx. MS/MS spectra for all DEX-2 studies have been uploaded into PeptideAtlas under dataset identifier PASS00302; http://www.peptideatlas.org/PASS/PASS00302.
References
- 1.Li N, Overkleeft HS, Florea BI. Curr. Opin. Chem. Biol. 2012;16:227–233. doi: 10.1016/j.cbpa.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 2.a) Speers AE, Cravatt BF. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]; b) Hang HC, Loureiro J, Spooner E, van der Velden AWM, Kim Y-M, Pollington AM, Maehr R, Starnbach MN, Ploegh HL. ACS Chem. Biol. 2006;1:713–723. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 3.Yang P-Y, Liu K, Zhang C, Chen GYJ, Shen Y, Ngai MH, Lear MJ, Yao SQ. Chem.-Asian J. 2011;6:2762–2775. doi: 10.1002/asia.201100306. [DOI] [PubMed] [Google Scholar]
- 4.Lee YH, Tan HT, Chung MCM. Proteomics. 2010;10:3935–3956. doi: 10.1002/pmic.201000289. [DOI] [PubMed] [Google Scholar]
- 5.a) Hillaert U, Verdoes M, Florea BI, Saragliadis A, Habets KLL, Kuiper J, Van Calenbergh S, Ossendorp F, van der Marel GA, Driessen C, Overkleeft HS. Angew. Chem. 2009;121:1657–1660. doi: 10.1002/anie.200805529. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:1629–1632. doi: 10.1002/anie.200805529. [DOI] [PubMed] [Google Scholar]; b) Fan F, Nie S, Dammer EB, Duong DM, Pan D, Ping L, Zhai L, Wu J, Hong X, Qin L, Xu P, Zhang Y-H. J. Prot. Res. 2012;11:5763–5772. doi: 10.1021/pr300575u. [DOI] [PubMed] [Google Scholar]
- 6.a) Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Mol. Cell. Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]; b) Kallemeijn WW, Li K-Y, Witte MD, Marques ARA, Aten J, Scheij S, Jiang J, Willems LI, Voorn-Brouwer TM, van Roomen CPAA, Ottenhoff R, Boot RG, van den Elst H, Walvoort MTC, Florea BI, Codée JDC, van der Marel GA, Aerts JMFG, Overkleeft HS. Angew. Chem. 2012;124:12697–12701. doi: 10.1002/anie.201207771. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2012;51:12529–12533. doi: 10.1002/anie.201207771. [DOI] [PubMed] [Google Scholar]
- 7.a) Kornhuber J, Tripal P, Reichel M, Mühle C, Rhein C, Muehlbacher M, Groemer TW, Gulbins E. Cell. Physiol. Biochem. 2010;26:9–20. doi: 10.1159/000315101. [DOI] [PubMed] [Google Scholar]; b) Kurishita Y, Kohira T, Ojida A, Hamachi I. J. Am. Chem. Soc. 2012;134:18779–18789. doi: 10.1021/ja308754g. [DOI] [PubMed] [Google Scholar]
- 8.Lübke T, Lobel P, Sleat DE. Biochim. Biophys. Acta, Mol. Cell Res. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Duve C, De Barsy T, Poole B, Trouet A, Tulkens P, Van Hoof F. Biochem. Pharmacol. 1974;23:2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 10.Anderson RG, Falck JR, Goldstein JL, Brown MS. Proc. Natl. Acad. Sci. U.S.A. 1984;81:4838–4842. doi: 10.1073/pnas.81.15.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Chem. Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 12.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis I. Biochem. Soc. Trans. 2009;37:1042–1044. doi: 10.1042/BST0371042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminskyy V, Zhivotovsky B. Biochim. Biophys. Acta, Proteins Proteomics. 2012;1824:44–50. doi: 10.1016/j.bbapap.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Levine B, Kroemer G. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Muno D, Sutoh N, Watanabe T, Uchiyama Y, Kominami E. Eur. J. Biochem. 1990;191:91–98. doi: 10.1111/j.1432-1033.1990.tb19097.x. [DOI] [PubMed] [Google Scholar]; b) Fuertes G, Martín De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ni H-M, Bockus A, Wozniak AL, Jones K, Weinman S, Yin X-M, Ding W-X. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn WA. J. Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dengjel J, Høyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, Farkas T, Kirkegaard T, Becker AC, Schroeder S, Vanselow K, Lundberg E, Nielsen MM, Kristensen AR, Akimov V, Bunkenborg J, Madeo F, Jäättelä M, Andersen JS. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristensen AR, Schandorff S, Høyer-Hansen M, Nielsen MO, Jäättelä M, Dengjel J, Andersen JS. Mol. Cell. Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Inubushi T, Shikiji M, Endo K, Kakegawa H, Kishino Y, Katunuma N. Biol. Chem. 1996;377:539–542. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.