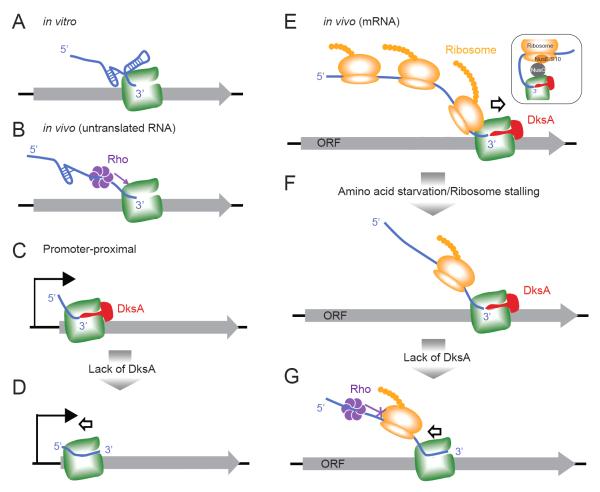
Figure 6. Model of opposing effects of DksA and stalled ribosome on transcription elongation.
(A, B) In the absence of translation in vivo or in vitro, RNA secondary structure prevents backtracking sterically (A). In vivo, Rho can be recruited to stalled ECs through the naked RNA and dislodge ECs (B). (C, D) During early transcription elongation at promoter-proximal regions, before significant upstream RNA structure forms to prevent backtracking, DksA modestly facilitates RNAP progression. (E) During in vivo mRNA transcription, the trailing ribosome prevents RNA/DNA backtracking on the ECs, by sequestering the mRNA, possibly aided by association of the ribosome and RNAP via NusG (inset; Burmann et al., 2010). (F-G) Upon amino acid starvation, translation is inhibited, allowing transcription stalling because uncoupling of transcription and translation frees unstructured segments of RNA that can backtrack through RNAP (the arrow indicates movement of RNAP relative to DNA; in vivo DNA must backtrack through a stationary RNAP that is constrained against rotation by the tethered ribosome). In this situation, DksA becomes crucial to maintain transcription elongation (F). In the absence of DksA, RNAP backtracks as it is no longer protected by the trailing ribosome or by RNA secondary structure, and unable to be released by Rho-dependent termination (G).