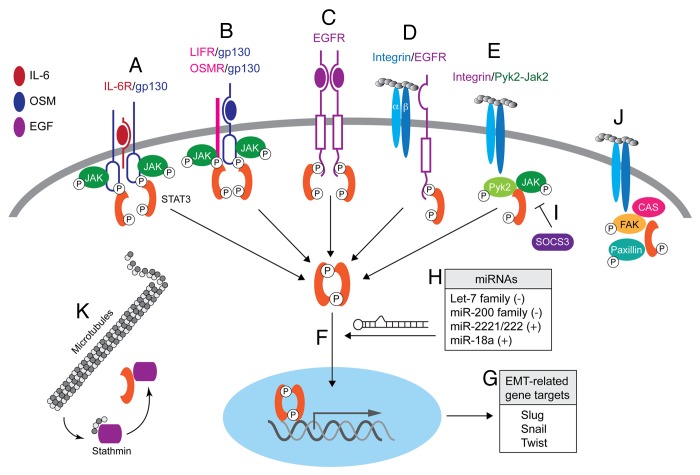
Figure 1. The differential modes of STAT3 activation and its genomic and nongenomic functions in responsive cells. (A) The IL-6/IL-6R complex induces dimerization of the common cytokine receptor subunit, gp130 leading the sequential activation of JAK2 and STAT3. (B) OSM binds gp130 and signals through LIF receptor (LIFR)/gp130 heterodimers, or through OSM receptor (OSMR)/gp130 heterodimers to activate JAK2-STAT3. (C) Ligand-activated EGFR forms a molecular complex with STAT3, leading to its activation. (D) FN adhesion induces formation of integrin-EGFR complexes and subsequent EGFR-dependent STAT3 activation. (E) FN-induced cell adhesion is also associated with STAT3 activation via an integrin:Pyk2:JAK2 molecular complex that functions independent of EGFR. (F and G) Tyrosine-phosphorylated STAT3 dimers undergo nuclear translocation (F) where they regulate several “master” EMT transcriptional networks (G). (H) Various miRs discussed in the text either restrict (−) or enhance (+) STAT3 signaling and EMT programs by targeting molecules associated with distinct aspects of the STAT3 pathway. (I) SOCS3 inhibits STAT3 signaling via blockade of upstream signaling through interactions with gp130 and JAK family members. (J) Tyrosine-phosphorylated STAT3 can localize to focal adhesions where it interacts with focal adhesion kinase (FAK) and paxillin, and also regulates the phosphorylation status of the adaptor protein p130Cas. (K) Cytosolic STAT3 also promotes microtubule polymerization by sequestering the microtubule-destabilizing factor stathmin.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.