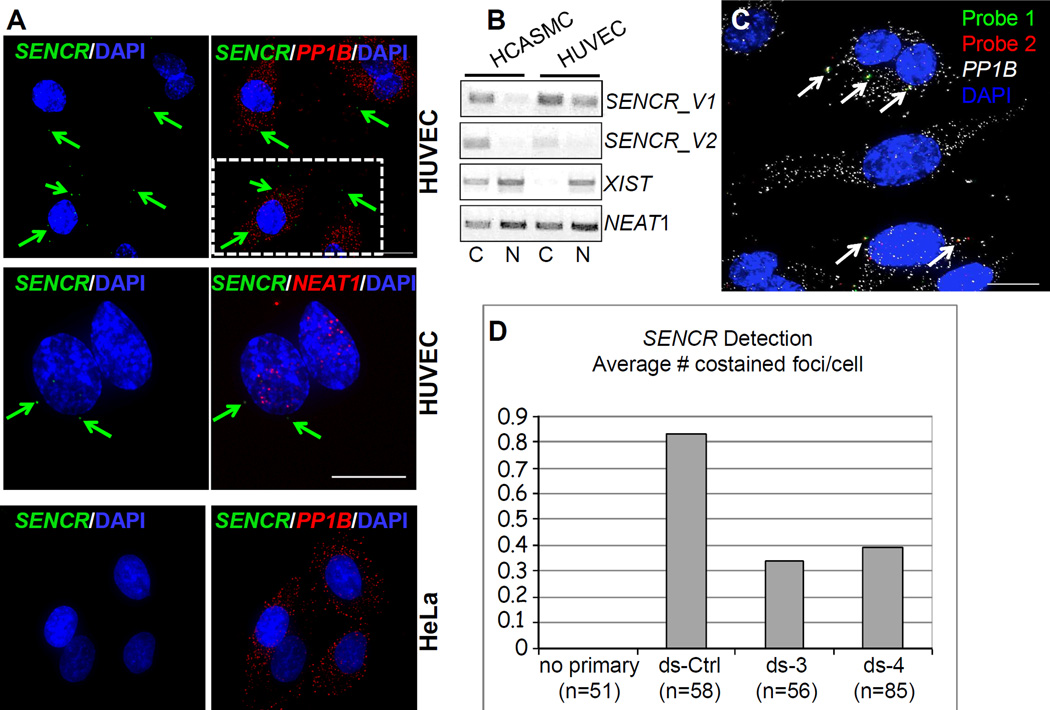
Figure 4. Localization of SENCR RNA.
(A) RNA FISH analysis of SENCR versus cytoplasmic PP1B mRNA and nuclear NEAT1 RNA in indicated cell types. Arrows point to single molecules of SENCR RNA in the cytoplasm of HUVEC. Scale bars are all 5 µm. The broken white rectangle at upper right is shown at higher magnification in Figure III in the online only Data supplement. (B) RT-PCR analysis of two SENCR isoforms versus other lncRNA genes from polyA+ RNA isolated from the cytoplasmic (C) or nuclear (N) fractions of indicated cell types. (C) Application of two probe sets to SENCR RNA (see Methods). Arrows point to coincident localization of two fluorescently tagged probe sets (yellow) targeting different regions of the SENCR transcript. (D) Quantitative measures of coincident localization of SENCR probes in HUVEC transfected with a dicer substrate control RNA (ds-Ctrl) or two dicer substrate RNAs targeting different regions of SENCR (ds-3 and ds-4). The Y-axis indicates the average number of co-stained foci/cell.