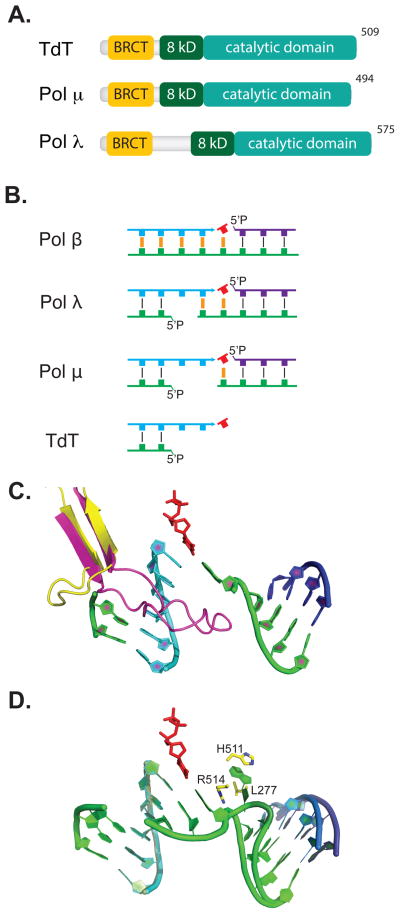
Fig. 4. Polymerases.
(A) Members of X family polymerases implicated in NHEJ possess a BRCA1 C-terminal (BRCT) domain, yellow, an 8 kilodalton (8kD), dark green, and a catalytic domain, teal. (B) The four mammalian Family X polymerases have activity with decreasing requirement for complementary sequence, orange, opposite primer, cyan, and incoming nucleotide triphosphate, red. Template strand, green, and downstream strand, purple, are also shown. (C) The loop1 motif in Pol λ, yellow, is shown from a structure of Pol λ bound to a 1-nucleotide gap with primer, cyan (1XSN) [153], downstream strand, purple, incoming nucleotide, red, and template, green. Four nucleotides of template opposite the primer were omitted. In place of these nucleotides we show loop1 from a structure of TdT, magenta (1JMS) [86], aligned with 1XSN. (D) Aligned substrates from structures of Pol λ with a 1 nt (1XSN) [153] versus a 2 nt gap (3HWT) [92]; primer, cyan, and incoming nucleotide, red, are super-imposable, as is most of template, green, and down stream strands, dark blue & purple. The extra template nucleotide is buried (“scrunched”) in a pocket formed by L277, H511, and R514.