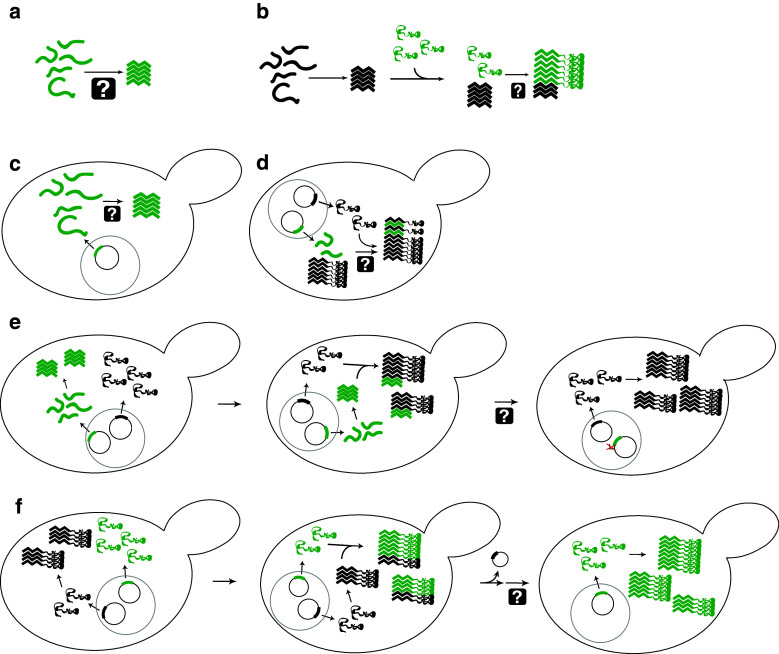
Fig. 2.
Assays to monitor prion-like activity and to define the regions of PFDs responsible for various aspects of prion activity. a In vitro aggregation. Protein fragments are incubated, generally with shaking, and aggregation is monitored using various techniques including Congo red binding, thioflavin T fluorescence, or pelleting assays. b Seeded in vitro aggregation. Preformed aggregates (green) are mixed with soluble protein (black) to test the ability of fragments to seed aggregation, or to test the ability of mutants to add onto preformed aggregates. c De novo aggregation in vivo. PFD fragments are transiently over-expressed. Aggregation is monitored either by fusing the fragments to GFP to observe foci formation, or through biochemical methods such as semi-denaturing detergent agarose gel electrophoresis (SDD–AGE). d Decoration of aggregates. PFD fragments are expressed in prion-positive cells to determine whether the fragments are capable of adding to preexisting aggregates. e Induction assays. PFD fragments (green) are transiently overexpressed in the presence of the full-length prion protein (black) to determine whether these fragments are sufficient to seed aggregation of the full-length protein. Aggregation is generally assayed by monitoring loss of function of the full-length protein. f Prion propagation assays. A prion-positive cell in which the chromosomal copy of the prion gene is deleted, but that carries a maintainer plasmid expressing the wild-type prion protein (black), is transformed with a plasmid expressing a prion protein mutant (green). The prion phenotype is assayed after selection for loss of the maintainer plasmid to determine if the mutant is capable of maintaining the prion