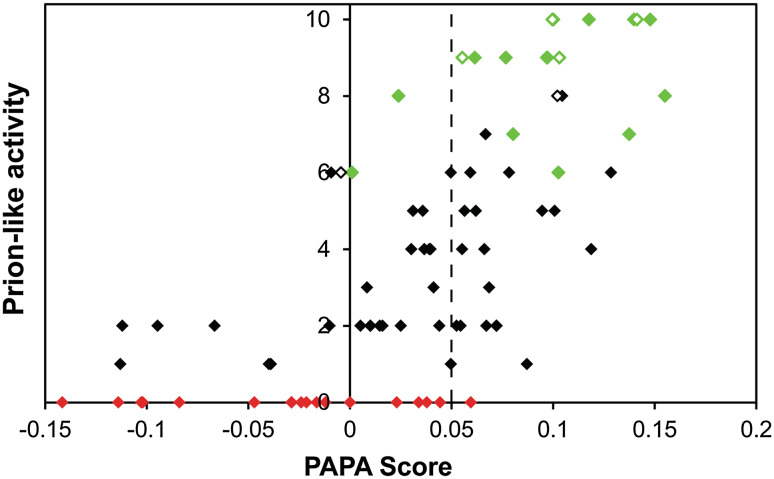
Fig. 3.
PAPA predictions for yeast prion-like proteins. Alberti et al. identified the 100 proteins with greatest compositional similarity to known yeast prions. Each was tested in four assays for prion-like activity, and given a prion-activity score from 0 to 10 based on these results. PAPA was then used to predict the prion-like activity of each protein. Domains that were not testable in one or more assays are excluded. Proteins that showed prion-like activity in all four assays are indicated in green. Proteins that failed to show prion-like activity in any assay are indicated in red. Known PFDs are indicated with open diamonds. The PAPA cutoff shown to most effectively discriminate between proteins with and without prion-like activity (0.05) is indicated with a dotted line. However, this is not an absolute cut-off; between 0 and 0.05, most proteins show at least some prion-like activity. Updated and adapted from [80]