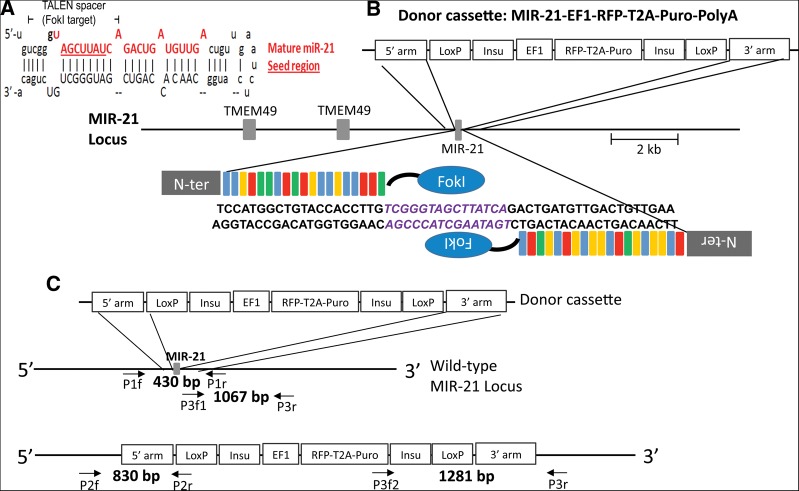
FIGURE 1.
Schematic overview of the targeting strategy against miR-21 in the human genome using TALENs in combination with an HR donor vector. (A) miR-21 stem–loop structure, the mature miR-21 sequence is shown in red; the seed region is underlined. TALENs were designed to position the miRNA seed region in the central portion of the spacer, directing cleavage to this functionally essential miRNA region. (B) A HR donor plasmid was created corresponding to the cleavage location of the TALEN pair and carried 509-bp (5′ arm) and 600-bp (3′ arm) regions of homology to the miR-21 sequence, which, in the native genome, are separated by 202 bp that include the miR21 stem–loop structure. Two LoxP sites are flanking the insulated expression cassette, which is composed of an EF1α promoter-driven RFP and puromycin-resistant gene (Puro), separated by a T2A linker (self-cleaving peptide sequence). (C) Locations of primers used for genotyping of HEK293 cells targeted with TALENs and HR donor. Primer pairs P2f, P2r and P3f, P3r were designed to amplify the junctions between genome and inserted HR donor cassette (830 bp and 1281 bp, respectively). Triple primers including two forward primers (P3f1 and P3f2) and one common reverse primer (P3r) were designed to co-amplify the HR knockout allele (P3f2 and P3r; 1281 bp) and the wild-type or NHEJ-modified allele (P3f1 and P3r; 1067 bp). Primer pair P1f and P1r was designed to amplify a 430-bp portion of the miR-21 region for subsequent sequence analysis to detect possible NHEJ events.