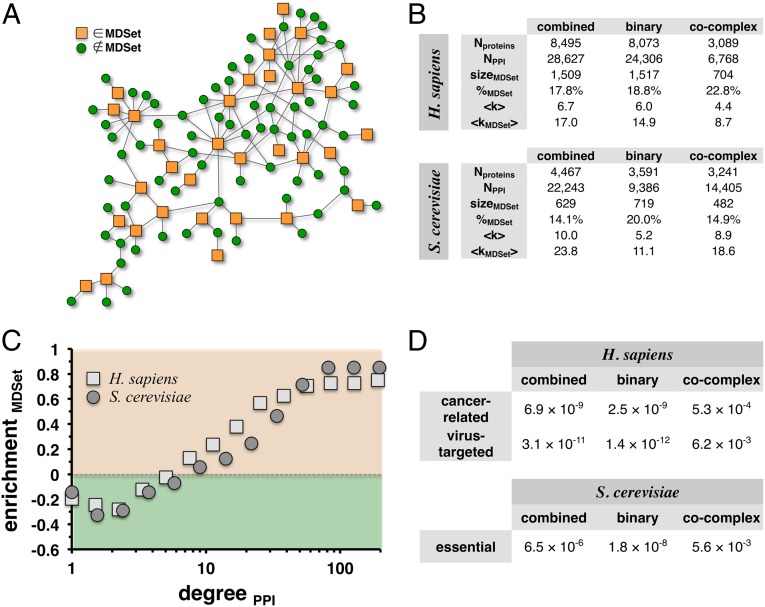
Fig. 1.
Definition and characteristics of minimum dominating sets. (A) In a toy network we illustrate the concept of an MDSet. Specifically, an MDSet is defined as an optimized subset of nodes (orange squares) from where each remaining (i.e., non-MDSet) node (green circles) can be immediately reached by one step. As a consequence, each non-MDSet protein is linked to at least one MDSet protein. (B) In the table we present statistics of protein interaction networks and their corresponding MDSets in human and yeast. In particular, we accounted for binary and cocomplex interactions as well as combined interaction datasets. (C) We grouped proteins in logarithmic bins according to their number of interaction partners in the combined networks. In each bin, we separately calculated the frequency of proteins in the full networks and MDSets. In comparison, MDSet proteins mostly appeared enriched in groups of proteins with roughly more than 10 interactions. (D) By applying a Fisher’s exact test, we found that cancer-related genes and proteins that are targeted by human viruses are significantly enriched in human MDSets. Similarly, essential genes are significantly present in the corresponding yeast MDSets.