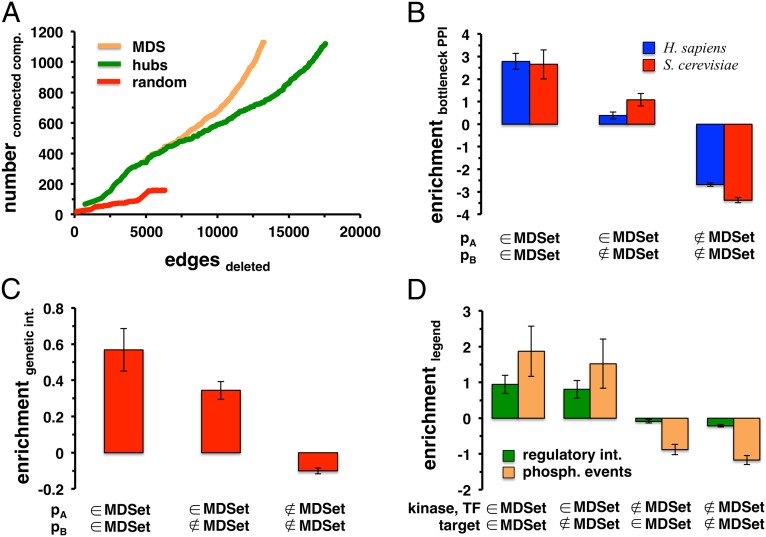
Fig. 2.
Enrichment of topological and functional entities in MDSets. In A we sorted all MDSet proteins in the combined yeast interaction network according to their degree. To provide an equivalent set of equal size we collected and sorted the highest connected hub proteins. Furthermore, we randomly sampled a set of yeast proteins of the same size. Starting with the most connected protein, we gradually deleted proteins and calculated the number of connected components in the altered network. In comparison, the deletion of MDSet proteins had a higher impact on the resilience of the underlying networks than hubs alone. (B) Defined as the top 10% of interactions with the highest edge betweenness, we determined a set of bottleneck interactions in the combined human and yeast networks, respectively, and counted their occurrence between (non)MDSet proteins (pA and pB). Randomly sampling MDSets 10,000 times, we observed that bottleneck interactions were significantly enriched between MDSet proteins and depleted between non-MDSet proteins (P < 10−4). In C we determined the enrichment of genetic interactions between yeast (non)MDSet proteins (pA and pB) in the combined network. Randomly sampling MDSet proteins 10,000 times we clearly observed that genetic interactions preferably appeared between MDSet proteins, whereas the opposite applied for non-MDSet proteins (P < 10−4). (D) Human phosphorylation events and transcriptional, regulatory interactions were significantly enriched when at least kinases or transcription factors were involved in the MDSet of the combined human network (P < 10−4).