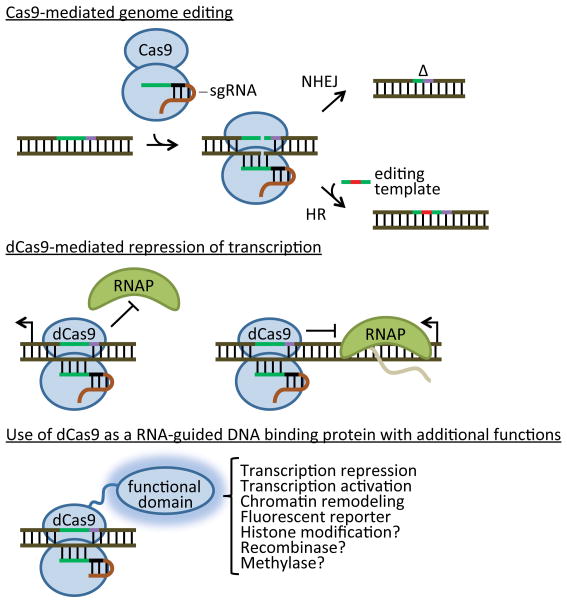
Figure 3. Cas9-based genetic applications.
Wild-Type Cas9 loaded with a single guide RNA (sgRNA) generates dsDNA breaks that can be used to introduce target mutations. Chromosomal breaks can be repaired by non-homologous end joining (NHEJ), creating indels (Δ) that introduce knock-out frameshift mutations. If a sequence homologous to the Cas9 target is provided (the editing template; either linear dsDNA or a short oligonucleotide), the break can be repaired by homologous recombination (Cong et al., 2013; Mali et al., 2013b). In this case, site-specific mutations in the editing template are can also be incorporated in the genome. A catalytically dead Cas9 (dCas9, containing mutations in both the RuvC and HNH actives sites) can be used as an RNA-guided DNA binding protein that can repress both transcription initiation when bound to promoter sequences or transcription elongation when bound to the template strand within an open reading frame (Bikard et al., 2013; Qi et al., 2013). dCas9 can also be fused to different functional domains to bring enzymatic activities and/or reporters to specific sites of the genome (Gilbert et al., 2013b).