FIG. 3.
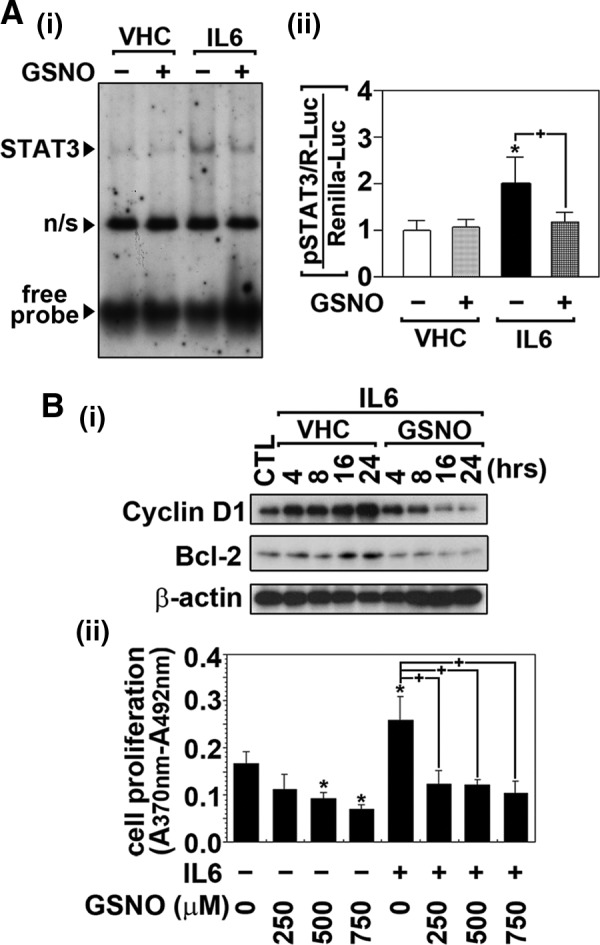
GSNO inhibits target DNA interaction of STAT3 and its transactivation by IL-6. (A) The effects of GSNO on IL-6-induced STAT3 target DNA binding activity and its transactivity were examined by gel-shift and reporter gene assays. For the gel-shift assay, BV2 microglia were pretreated with GSNO (500 μM) for 2 h and then treated with IL-6 (30 ng/mL) for 30 min. Nonspecific reaction is indicated by n/s (A-i). For the reporter gene assay, BV2 microglia transfected with STAT3-responsive luciferase construct (pSTAT3/R-Luc) and renilla luciferase contruct (phRL-CMV) as transfection control were pretreated with GSNO (500 μM) for 2 h and then treated with IL-6 (30 ng/mL) for 24 h. STAT3 transactivities were analyzed by luciferase activity assay as described under experimental procedure (A-ii). (B) The effects of GSNO on the expression of STAT3 downstream gene expression and cell proliferation were examined in BV2 microglia. The cells were treated with GSNO (500 μM) for 2 h and then treated with IL-6 (30 ng/mL) for indicated time points. The cellular levels of cyclin D1 and Bcl-2 were then analyzed by Western analysis (B-i). The effect of pretreatment of BV2 cells with increasing concentrations of GSNO for 2 h on IL-6-induced (30 ng/mL for 14 h) cell proliferation was analyzed by BrdU incorporation assay (B-ii). The vertical lines in A-ii and B-iii indicate the standard error of mean; *p<0.05 compared with vehicle-treated (VHC; dimethylsulfoxide or phosphate-buffered saline) control groups; +p<0.01 compared with IL-6-treated groups. BrdU, 5-bromo-2′-deoxyuridine.
