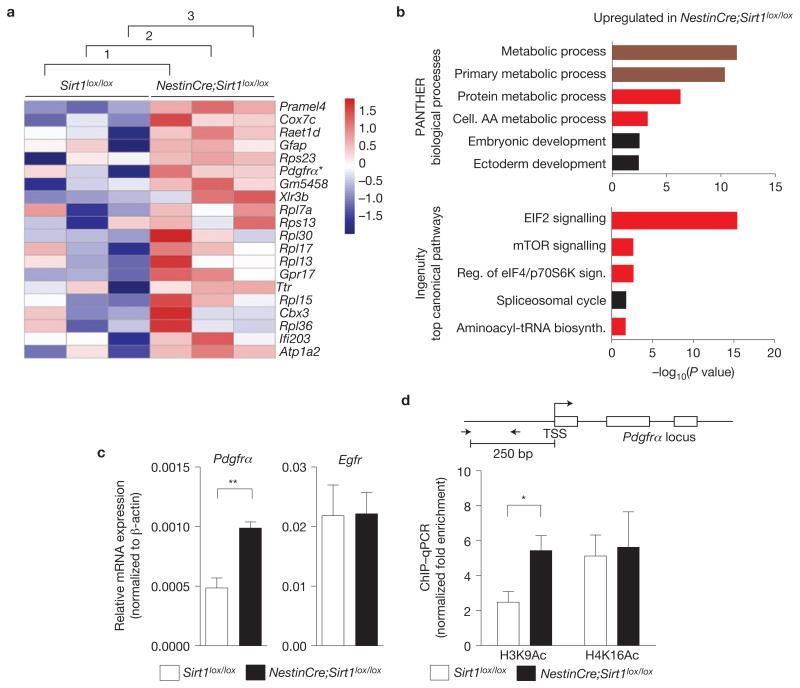
Figure 7.
SIRT1 inactivation leads to the upregulation of genes involved in metabolism, protein translation and growth factor signalling. (a) Genes upregulated in NestinCre;Sirt1lox/lox NSCs and neural progenitors compared with Sirt1lox/lox control NSCs and neural progenitors. Whole-genome microarray data from three independent biological replicates of proliferating NestinCre;Sirt1lox/lox and Sirt1lox/lox NSCs and neural progenitor isolated from 8-week-old mice. Heat map represents a subset of genes upregulated in the absence of active SIRT1 that was selected on the basis of a false discovery rate of less than 0.001%. Numbers 1–3 designate independent biological experiments. Colours represent the Z score of the robust multi-array analysis-normalized expression level for each gene (red is high expression; blue is low expression). Asterisk highlights Pdgfrα, a gene important for OPC proliferation and maintenance9,10,52,53. (b) Genes upregulated following SIRT1 inactivation are significantly over-represented in biological processes related to metabolism and ribosomal biology. Genes differentially regulated by SIRT1 were selected on the basis of a false discovery rate of less than 1% and analysed using PANTHER and Ingenuity Pathway Analysis (IPA). Canonical pathways significantly over-represented in the genes upregulated following SIRT1 inactivation are presented. Upper graph shows 6 of the top 7 PANTHER biological processes. Lower graph shows the top 5 IPA processes. Dark red colour highlights categories related to metabolism and bright red colour highlights categories related to protein synthesis and metabolism. AA, amino acid. (c) Pdgfrα mRNA expression is greater in proliferating NSCs and neural progenitors lacking active SIRT1 than in control cells. RT–qPCR on total RNA from primary NSC and neural progenitor cultures from NestinCre;Sirt1lox / lox and Sirt1lox/lox 8-week-old mice with primers to Pdgfrα and Egfr. Expression of Pdgfrα and Egfr was normalized to β-actin. Mean±s.e.m. of 3 independent experiments. Two-tailed, unpaired Student’s t-test, **P = 0.0068. (d) Increased H3K9 acetylation at the Pdgfrα promoter in NSCs and neural progenitors lacking active SIRT1. ChIP-qPCR on NSCs and neural progenitors from Sirt1lox / lox and NestinCre;Sirt1lox/lox 8-week-old mice with antibodies against H3K9Ac or H4K16Ac with primers to the Pdgfrα promoter. Data are presented as fold enrichment at the Pdgfrα promoter normalized to that at the promoter of HoxA10, a silent gene in these cells. Mean±s.e.m. of 3 independent experiments. Two-tailed, unpaired Student’s t-test, *P = 0.0491. TSS, transcriptional start site. The Pdgfrα locus is not drawn to scale.