Figure 4.
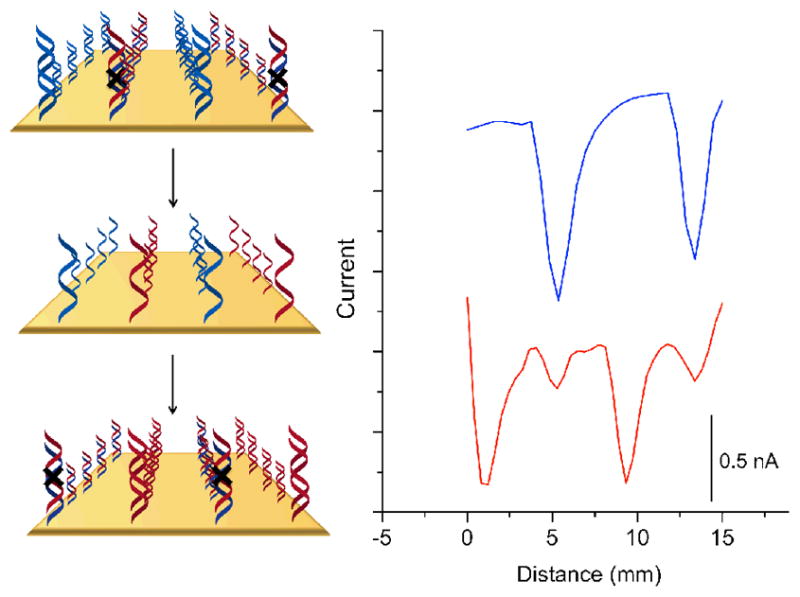
Oligonucleotide detection through dehybridization and hybridization. Current increases negatively down the y-axis. The surface was patterned with two strips of well-matched DNA and two strips that contain a mismatch. The blue trace is a preliminary scan in 2μM methylene blue and 200μM ferricyanide before dehybridization. The surface was subsequently soaked in phosphate buffer (5 mM phosphate, 50 mM NaCl, pH 7) at 65°C for 15 minutes. Single stranded oligonucleotides complementary to the formerly mismatched sequence heated to 65°C were added and allowed to cool to room temperature over 1 hour. The red trace shows the post-rehybridization data, where the mismatched sequences are now well matched and the formerly well matched, now mismatched.
