Abstract
The gene-silencing activity of a small interfering RNA (siRNA) is determined by various factors. Considering that RNA interference (RNAi) is an unparalleled technology in both basic research and therapeutic applications, thorough understanding of the factors determining RNAi activity is critical. This report presents observations that siRNAs targeting KRT7 show cell-line-dependent activity, which correlates with the expression level of KRT7 mRNA. By modulating the target mRNA level, it was confirmed that highly expressed genes are more susceptible to siRNA-mediated gene silencing. Finally, several genes that show different expression levels in a cell-line dependent manner were tested, which verified the expression-level-dependent siRNA activities. These results strongly suggest that the abundance of target mRNA is a critical factor that determines the efficiency of the siRNA-mediated gene silencing in a given cellular context. This report should provide practical guidelines for designing RNAi experiments and for selecting targetable genes in RNAi therapeutics studies.
Introduction
Small interfering RNA (siRNA) is an effector molecule of RNA interference (RNAi), and consists of an RNA duplex with length between 19 and 21 bases (Elbashir et al., 2001a, 2001b). Once introduced into a cell, siRNA induces target messenger RNA (mRNA) degradation in a sequence-specific manner via the RNAi pathway, resulting in the inhibition of target gene expression (Carthew and Sontheimer, 2009). In general, potent siRNAs have IC50 values in the sub-nanomolar range, and treatment with 1–10 nM of siRNA with the aid of a delivery vehicle is sufficient to achieve effective target gene silencing in a cell culture model (Kretschmer-Kazemi Far and Sczakiel, 2003). Accordingly, siRNA-mediated gene knockdown technique is regarded as a most powerful tool for in vitro gene function study, and has a wide range of application, from single gene analysis to genome-wide screening studies (Dorsett and Tuschl, 2004).
Although there are many aspects to consider in designing an experiment using siRNA, it is most important that the siRNA used in the experimental model has potent gene-silencing activity. For this reason, there have been many efforts to identify features associated with the gene silencing potency of siRNA since the beginning of siRNA technology development. A series of reports identified features and algorithms related to the siRNA activity, which helps to design siRNAs with good gene silencing potency to some extent (Elbashir et al., 2002; Reynolds et al., 2004; Ui-Tei et al., 2004; Gong et al., 2006, 2008; Matveeva et al., 2007; Lu and Mathews, 2008; Naito and Ui-Tei, 2012). Nevertheless, many studies report the observation of insufficient knockdown in siRNA experiments. For instance, a large-scale siRNA experiment reported a group of non-targetable genes even after testing repeated redesigns of the siRNA sequences (Krueger et al., 2007). This gene-specific response to siRNA treatment may be due to the inherent property of the transcript. However, identification of the underlying factors is still under investigation.
It is thought that a number of distinct features are linked to the siRNA efficacy, and they often work in a combined manner. These include properties of the siRNA sequence such as internal stability, as well as features of the target mRNA such as the secondary structure surrounding the siRNA target site (Bohula et al., 2003; Khvorova et al., 2003; Kretschmer-Kazemi Far and Sczakiel, 2003; Overhoff et al., 2005). In addition to the factors restricted to the interface of siRNA and target mRNA interaction, features that influence behavior in an indirect manner have been explored by several groups. It was suggested that cellular localization and the RNA-associated protein binding of the target mRNA may affect siRNA efficacy (Holen et al., 2002). Recently, competition between off-targets and on-targets were investigated, and it was reported that the presence of a large number of off-targets in the siRNA treated cells renders a “dilution effect” that limits siRNA activity on on-target mRNA (Arvey et al., 2010). Correlation between siRNA efficacy and target mRNA turnover rate has also been examined, and it was suggested that short-lived mRNAs are more insensitive to siRNA treatment (Larsson et al., 2010). In addition, an abundance of target transcript was proposed as one of the features affecting siRNA efficacy, and Xiuyuan Hu et al. clearly showed that transcripts with low expression levels are less susceptible to siRNA-mediated degradation through the analysis of RNAi-mediated knockdown efficiencies of sense and antisense transcript from the same genetic locus (Hu et al., 2004).
In this study, an RNAi experiment was designed for the functional study of the KRT7 gene. Unexpectedly, two independent KRT7 siRNAs showed dramatic differences in efficacy between two groups of cell lines. Further analysis revealed that the two groups of cell lines have different levels of KRT7 mRNA. Based on this initial finding, correlation between siRNA efficacy and target mRNA abundance was investigated with an inducible gene expression model and additional examples of the endogenous genes. It is proposed that the abundance of target mRNA is one of the key factors that determine the gene-silencing efficiency of siRNA in a given cellular context.
Materials and Methods
siRNAs
Chemically synthesized RNAs were purchased from Bioneer and annealed according to the manufacturer's protocol. The RNA sequences for each gene and additional information are provided in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/nat).
Cell culture and siRNA transfection
A549 (CCL-185), HEK293 (CRL-1573), HeLa (CCL-2), and SK-N-SH (HTB-11) cell lines were purchased from ATCC and maintained at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum (Gibco). Cells were routinely subcultured to maintain exponential growth. For transfection, cells were plated on 12-well plates 24 hours before transfection at 30%–50 % confluence in complete medium. Lipofectamine RNAiMAX was used for siRNA transfection following the manufacturer's protocol (Invitrogen). The final concentrations of siRNA transfected were 1 and 10 nM for KRT7 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and 10 nM for other genes. Cells were harvested 24 hours after siRNA transfection.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from cells using an Isol-RNA Lysis Reagent (5 Prime) according to manufacturer's instructions. Total RNA (500 ng) was used as a template for complementary DNA (cDNA) synthesis using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Aliquots (1/20) of each cDNA reaction were analyzed by quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR) using the StepOne Real-Time PCR System (Applied Biosystems). Gene-specific primers were mixed with SYBR Premix Ex Taq (Takara). In the analysis of the siRNA target gene knockdown efficiency (Figs. 1A, B, 2C, and 3B), the target genes and TUB1A1 (internal control) mRNA levels were determined using the relative standard curve quantitation method. Data from a standard dilution series of the control sample were used to generate the standard curve. For the cDNA copy number analysis (Figs. 1C, 3A), the standard curve quantitation method was used. The PCR product of each gene was purified using a Qiaquick PCR purification kit (Qiagen), and a series of diluted PCR products was used for generating the standard curve (Supplementary Fig. S1). The primer sequences for each gene and additional information are provided in Supplementary Table S2.
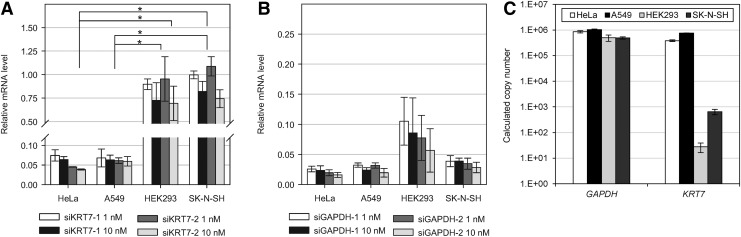
FIG. 1.
Analysis of KRT7 and GAPDH small interfering RNAs (siRNAs) activities in four different cell lines. (A) Activities of two siRNAs targeting KRT7 messenger RNA (mRNA). (B) Activities of two siRNAs targeting GAPDH mRNA. For A and B, siRNAs were transfected into each cell lines and target mRNA levels were analyzed by quantitative real-time reverse transcription polymerase chain reaction (PCR) 24 hours following transfection. The values plotted as “relative mRNA levels” on the y-axis in the figure were calculated as the target mRNA level divided by the TUBA1A (control) mRNA level and relative mRNA levels to mock transfection control (reagent only) are presented. All data in the graph represent mean±SD values of three independent experiments. Student's t-test was used to compare target mRNA knockdown efficiency between two cell lines. *P<0.01. (C) Expression level of KRT7 and GAPDH in four cell lines. Copy number of KRT7 or GAPDH cDNAs in reaction was analyzed through quantitative real-time PCR. Serial dilutions of purified PCR product of each gene were used for generating standard curve. All data in the graph represent mean±SD values of three independent samples.
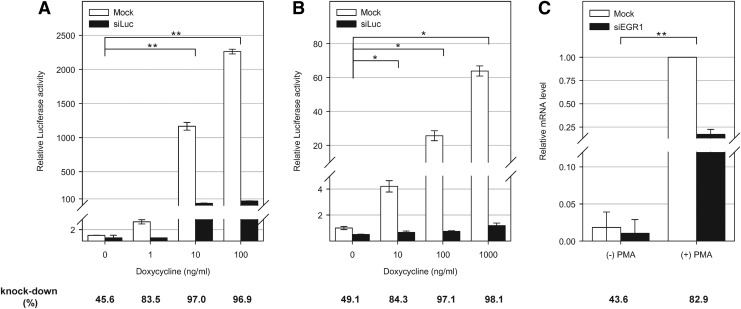
FIG. 2.
Analysis of luciferase and EGR1 (early growth response 1) siRNAs activities in non-induction or induction conditions. (A) Analysis of luciferase siRNA activities in HEK293/t3g-9 cell line. HEK293/t3g-9 cells were co-transfected with pTRE3G-Luc and pRL-SV40 and the indicated concentration of doxycycline (0–100 ng/mL) was applied. Twenty-four hours later, luciferase activity was measured. (B) Analysis of luciferase siRNA activities in HeLa/t3g-luc-24 cell line. Indicated concentration of doxycycline (0–1000 ng/mL) was treated for 24 hours, and luciferase activity was measured. (A–B) For knockdown study, siLuc (10 nM final) is transfected for 24 hours and relative luciferase activities to no doxycycline condition were presented as bar graphs. Three independent experiments were performed in triplicate manner, and data in the graph represent mean±SD values of one representative experiment. (C) Analysis of siEGR1 activities in A549 cells under with or without phorbol-12-myristate-13-acetate (PMA) stimulation. A549 cells are transfected with siEGR1 under with or without PMA stimulation (100 nM, 24 hours) and target mRNA levels were analyzed by quantitative real-time reverse transcription PCR as described in Fig. 1A. Relative mRNA levels to PMA stimulation and mock transfection condition are presented. All data in the graph represent mean±SD values of three independent experiments. (A–C) Average percent knockdown efficiencies are indicated at the bottom of each condition. Student's t-test was used to compare target mRNA knockdown efficiency between non-induction and induction conditions. *P<0.01; **P<0.05.
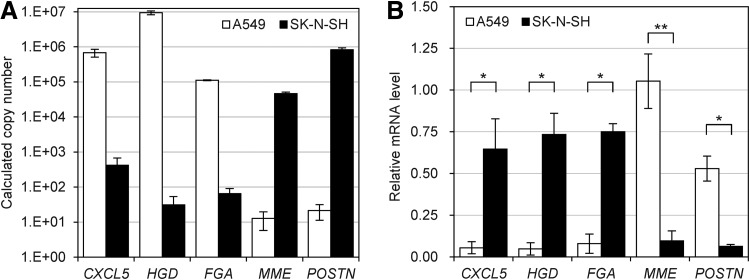
FIG. 3.
Analysis of cell line specific siRNA activity. (A) Expression level of five genes in A549 and SK-N-SH cell lines. Copy number of each gene cDNA was analyzed and presented as described in Fig. 1B. (B) Activities of five siRNAs in A549 and SK-N-SH cell lines. siRNAs targeting each gene were transfected and target mRNA levels were analyzed as described in Fig. 1A. All data in the graphs represent mean±SD values of three independent samples. Student's t-test was used to compare target mRNA knockdown efficiency between two cell lines. *P<0.01; **P<0.05.
Establishing stable cell lines
Tet-On 3G Tetracycline Inducible Gene Expression Systems (Clontech) were used for establishing cell lines according to the manufacturer's protocol. Briefly, for establishing stable cell lines expressing Tet-On 3G, pCMV-Tet3G was transfected into HEK293 or HeLa cells using Xfect transfection reagent. G-418 resistant single colonies were picked after 2 weeks of G-418 selection (800 μg/mL for HEK293 and 400 μg/mL for HeLa, Gold Biotechnology) and tested for luciferase induction. The resulting HEK293/t3g-9 and HeLa/t3g-9 cell lines were maintained in G-418-containing media (400 μg/mL for HEK293 and 200 μg/mL for HeLa). For establishing a double-stable cell line, pTRE3G-Luc and hygromycin linear selection markers were cotransfected into HeLa/t3g-9 cells using Xfect. Hygromycin-resistant single colonies were picked after 2 weeks of hygromycin selection (200 μg/mL, Sigma-Aldrich) and tested for luciferase induction. The resulting HeLa/t3g-luc-24 was maintained in hygromycin-containing media (100 μg/mL).
Luciferase knockdown analysis of HEK293/t3g-9 cell line
Cells were plated on 24-well plates at 50% confluence in complete medium. After 24 hours, pTRE3G-Luc (100 ng) and pRL-SV40 (1 ng, Promega) were cotransfected using Lipofectamine 2000 reagent following the manufacturer's protocol (Invitrogen). For knockdown study, siLuc-targeting luciferase (10 nM final) is included in the transfection mixture. After transfection, doxycycline (Clontech) was added at the indicated concentration for 24 hours, and luciferase activity was analyzed using the Dual Luciferase Reporter Assay System (Promega). Results were quantified with the 20/20n Luminometer (Turner Biosystems).
Luciferase knockdown analysis of HeLa/t3g-luc-24 cell line
Cells were plated on 24-well plates at 50% confluence in complete medium. After 24 hours, siLuc (10 nM final) was transfected using Lipofectamine RNAiMAX according to the manufacturer's protocol (Invitrogen). After transfection, doxycycline was added at the indicated concentration for 24 hours. Luciferase activity was analyzed using the Luciferase Assay System (Promega), and the results were quantified with the 20/20n Luminometer.
Statistical analysis
Statistical significance was assessed by Student's t-test.
Results and Discussion
To knock down KRT7 gene expression, two siRNAs (siKRT7-1 and siKRT7-2) targeting the KRT7 mRNA sequence were designed. The transfection of each siRNA into the HeLa cell line showed efficient reduction of the target mRNA level at concentrations generally used for RNAi experiments (1 nM and 10 nM). Figure 1A shows that 1 nM transfection of both siKRT7s reduced ∼90% of the target mRNA level. Additional cell lines derived from different human tissues were then chosen, including A549, HEK293, and SK-N-SH, and RNAi experiments were conducted using siKRT7s. The treatment of siKRT7s into A549, a lung carcinoma cell line, resulted in similar inhibition of the target mRNA level. As shown in Fig. 1A, more than 90% knockdown of the target gene was achieved at 1 nM transfection using both siRNAs.
However, unlike HeLa and A549 cell lines, significant down-regulation of KRT7 mRNA could be not observed using siKRT7s in HEK293 and SK-N-SH cell lines. Seventy percent of the target mRNA remained under 10 nM siKRT7 transfection conditions For HEK293, a human embryonic kidney cell line. For SK-N-SH, a neuroblastoma cell line, the treatment of both siKRT7s achieved merely 20% reduction of the target mRNA level (Fig. 1A).
Among the factors affecting the efficacy siRNA-mediated target gene inhibition, the efficiency of siRNA delivery or the capacity of RNAi machinery might have a role in cell-line-dependent siRNA activity. Particularly, some cell types such as human T cell lines are known to have difficulty achieving high transfection efficiency (Ovcharenko et al., 2005). However, all four cell lines used in this study are widely used for RNAi experiments, and multiple reports have described efficient target gene knockdown by RNAi methods in these cell lines (Elbashir et al., 2001a; Chiu and Rana, 2002; Truss et al., 2005; Ren et al., 2009).
We checked transfection efficiency of the four cell lines through the quantification of delivered siRNAs and the results showed similar level of transfected siRNAs in four cell lines (Supplementary Fig. S2A). Fluorescence visualization of the Cy3-labeled siRNA after transfection further confirmed the similar transfection efficiency of the siRNA molecules (Supplementary Fig. S2B). In addition, an RNAi experiment using two siRNAs targeting the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene (siGAPDH-1 and siGAPDH-2) showed potent RNAi activity in both HEK293 and SK-N-SH cell lines. As shown in Fig. 1B, siGAPDHs showed comparable target gene silencing in SK-N-SH cell lines compared to either HeLa or A549 cell lines. Up to 90 % reduction of the GAPDH mRNA level in the HEK293 cell line was also observed. These results confirm that the differences in RNAi activity among different cell lines cannot explain differences in KRT7 RNAi efficiency among the cell lines tested.
Larsson et al. investigated the relationship between the efficacy of siRNA and the turnover rate of mRNA targets. They showed that genes with high turnover rates are more resistant to siRNA-mediated silencing (Larsson et al., 2010). However, examination of the KRT7 mRNA half-life in four cell lines revealed that KRT7 mRNA in HeLa cell has the highest turnover rate among the cell lines tested. The half-life of KRT7 mRNA is 7 hours in HeLa cells; it is more than 24 hours in other cell lines (data not shown). Thus, the turnover rate factor also cannot explain the cell-line-dependent KRT7 siRNA efficacy observed.
Interestingly, real-time PCR raw data showed significant differences in cycle threshold (CT) values between siKRT7-responsive cell lines (HeLa and A549) and nonresponsive cell lines (HEK293 and SK-N-SH). In the experimental conditions, the CT values of KRT7 for HeLa and A549 were 18.1 and 16.7, respectively. For the HEK293 and SK-N-SH cell lines, KRT7 showed CT values of 30.1 and 27.6, respectively (For no-template control, PCR amplification signal was not detected and CT value was not determined). On the other hand, all tested cell lines have similar CT values of GAPDH, ranging from 15 to 16.
In general, the CT value in real-time PCR data directly reflects the amount of PCR targets, so the findings suggest that cell-line-specific siRNA activity correlates with the abundance of target mRNA level in the cell line tested. The copy number of each cDNA in reverse transcribed samples was analyzed, and the results revealed that HeLa and A549 cells contain a much higher number of KRT7 transcripts than SK-N-SH and HEK293 cells with a difference of three orders of magnitude (Fig. 1C).
The experimental observations using KRT7 siRNAs led to the examination of the correlation of target mRNA abundance and siRNA efficacy. To provide identical experimental conditions except for the target mRNA abundance, an experimental model that uses a tetracycline-inducible expression system was set up (Gossen and Bujard, 1992). In this system, the transcription of a gene of interest can be regulated by an induction molecule (doxycycline) in a dose-dependent manner. HEK293/t3g-9 cell line stably expressing Tet-On 3G Transactivator Protein was established. The transient transfection of pTRE3G-Luc vector along with doxycycline treatment caused more than a 2,000-fold difference of luciferase activity in HEK293/t3g-9 cells (Fig. 2A). siRNA targeting luciferase (siLuc) and pTRE3G-Luc vector were cotransfected in either non-induction or induction conditions. Reduced luciferase activity resulting from the siLuc-mediated target mRNA knockdown was analyzed. The results showed the correlation of siRNA efficacy and target abundance. As shown in Fig. 2A, knockdown level of luciferase activity by siLuc varied from 45.6% to 96.9% as the target gene expression level increased.
Further tests were conducted using a double stable cell line, HeLa/t3g-luc-24, which expresses Tet-On 3G activator protein and luciferase under the control of a PTRE3G promoter. The doxycycline-dose-dependent induction of luciferase activity in the cell line was confirmed, and an influence of the target gene level on siRNA efficacy was observed. In non-induction conditions, siLuc transfection resulted in 49.1% reduction of luciferase activity. On the other hand, 1000 ng/ml of doxycycline treatment resulted in 60-fold induction of luciferase activity, and siLuc treatment showed 98.1% reduction of luciferase activity in these conditions (Fig. 2B).
A model using an endogenously expressed gene was also tested. Early growth response protein 1(EGR1) is a stress-responsive gene, and the expression of EGR1 is induced by various stimulations (Adamson and Mercola, 2002; Thiel and Cibelli, 2002). The induction of EGR1 mRNA expression under phorbol-12-myristate-13-acetate (PMA) treatment conditions was tested in A549 cells. As shown in Fig. 2C, EGR1 showed low transcript level in basal conditions, and PMA treatment for 24 hours resulted in a 54-fold increase of the EGR1 mRNA level. siRNA targeting EGR1 was then transfected with or without PMA treatment conditions, and the siEGR1 target gene knockdown efficiency was analyzed. Similar to the doxycycline induction model experiment, it was confirmed that siERG1 showed target gene expression level-dependent efficiency. In non-inducing conditions, about 56.4% of the target mRNA remained 24 hours after siRNA treatment. On the other hand, siEGR1 treatment under PMA stimulation reduced the target mRNA level to 17.1%.
As a next step, additional genes that show differential expression levels between two cell lines were examined. The genome-wide transcript level of RNA in A549 and SK-N-SH cell lines was obtained from The Encyclopedia of DNA Elements (ENCODE) project RNA-seq data (Rosenbloom et al., 2012), and 5 genes showing distinct transcript levels between A549 and SK-N-SH cell lines were selected. CXCL5, HGD, and FGA are referred to as the “A549 genes” because they show high expression levels in the A549 cell line but low expression levels in the SK-N-SH cell line. The “SK-N-SH genes,” MME and POSTN, have an opposite expression pattern in the two cell lines (Fig. 3A). siRNAs targeting each gene were designed, and the target knock down efficiencies of individual siRNAs in the two cell lines were examined.
As expected, the results showed cell-line-dependent siRNA activities in accordance with the expression levels. As shown in Fig. 3B, the A549 genes, CXCL5, HGD, and FGA, showed efficient gene knockdown ranging from 92.1% to 95.2% in the A549 cell line. The same was true for the SK-N-SH genes in the SK-N-SH cell line (the knockdown efficiencies of MME and POSTN are 90.3% and 93.7%, respectively). In contrast, the transfection of the siRNAs targeting A549 genes in SK-N-SH cells and the siRNAs targeting SK-N-SH genes in A549 cells merely achieved 47% knockdown at most (Fig. 3B). The results further support the hypothesis that the resistance of a gene to siRNA treatment is frequently caused by low transcript level.
In the present study, dramatic differences in KRT7 siRNA efficacies between two groups of the cell lines led us to survey the correlation between siRNA efficacy and target mRNA abundance. Experimental model systems were then established to examine the relationship. In the same cellular context, target gene expression was manipulated and siRNA efficacy was analyzed. The results from the model systems using both exogenous (luciferase) and endogenous (EGR1) genes support the hypothesis that a low abundance of target mRNA leads to an inefficient knockdown by siRNA treatment. Five additional genes that show distinct expression levels in A549 and SK-N-SH cells were selected and the results showed siRNA efficacies that were dependent on the gene expression level.
Previously, Xiuyuan Hu et al. analyzed the correlation between the target transcript and siRNA efficacy and provided evidence that low-abundant transcripts are less susceptible to siRNA-mediated degradation. (Hu et al., 2004). However, to our knowledge, detailed, quantitative criteria of target transcript abundance that could be a practical guideline for siRNA experiments have not been provided. In this study, the correlation between target mRNA abundance and siRNA efficacy was explored by testing nine genes, and the results provide the range of non-targetable transcript level. In the experimental conditions, genes with cDNA numbers from 13 (MME in A549) to 420 (CXCL5 in SK-N-SH) were generally non-targetable by siRNA treatment. In contrast, genes with cDNA numbers higher than about 46,440 (MME in SK-N-SH) were readily targeted by siRNA.
More recently, the influence of gene expression level on siRNA silencing potential was explored through a large-scale siRNA validation screening (Krueger et al., 2007). The ΔCT values between the CT values of the target gene and GAPDH gene were used for indicators of the target gene abundance. Krueger et al., observed efficient knockdown in a set of genes with ΔCT>10, demonstrating that a low abundance of target gene does not have an effect on siRNA potency. In the present study, siEGR1 study showed similar results. Under PMA treatment conditions, ΔCT of EGR1 to GAPDH was 11.8, and efficient target gene knockdown was observed. However, under un-stimulated conditions with ΔCT of 16.5, similar knockdown efficiency was not achieved by siEGR1 treatment (Fig. 2C). The result suggests that genes with ΔCT to GAPDH ranging from 10 to 12 have sufficient number of transcripts to show efficient knockdown by siRNA treatment. CT values of the tested genes were also inspected, as shown in Fig. 3, which showed that all non-targetable transcripts have ΔCTs higher than 16. This result indicates that ΔCT higher than 16 to GAPDH may serve as an indicator of potential non-targetability in siRNA experiment.
The assessment of the target gene transcript level is a relatively straightforward process, and the data suggests that the evaluation of target gene abundance is an indispensable step in designing siRNA experiments. The validation of a sufficient target gene level prior to conducting siRNA experiments is expected to increase the success rate of target gene silencing, and by providing information for selecting an adequate experimental model, it may prevent misinterpretation of data from RNAi experiments. It is also believed that the inspection of target gene abundance is a useful process to exclude false positives in large-scale siRNA experiments such as genome-wide RNAi screening.
Supplementary Material
Acknowledgments
This work was supported by a Global Research Laboratory grant from the Ministry of Education, Science, and Technology (MEST) of Korea (no. 2008-00582). SWH was supported by a grant from the National Research Foundation of Korea funded by the Korean Government (Ministry of Education, Science and Technology) [NRF-2010-359-E00007].
Author Disclosure Statement
No competing financial interests exist.
References
- ADAMSON E.D., and MERCOLA D. (2002). Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol. 23, 93–102 [DOI] [PubMed] [Google Scholar]
- ARVEY A., LARSSON E., SANDER C., LESLIE C.S., and MARKS D.S. (2010). Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 6, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOHULA E.A., SALISBURY A.J., SOHAIL M., PLAYFORD M.P., RIEDEMANN J., SOUTHERN E.M., and MACAULAY V.M. (2003). The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem. 278, 15991–15997 [DOI] [PubMed] [Google Scholar]
- CARTHEW R.W., and SONTHEIMER E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIU Y.L., and RANA T.M. (2002). RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 10, 549–561 [DOI] [PubMed] [Google Scholar]
- DORSETT Y., and TUSCHL T. (2004). siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3, 318–329 [DOI] [PubMed] [Google Scholar]
- ELBASHIR S.M., HARBORTH J., LENDECKEL W., YALCIN A., WEBER K., and TUSCHL T. (2001a). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- ELBASHIR S.M., HARBORTH J., WEBER K., and TUSCHL T. (2002). Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199–213 [DOI] [PubMed] [Google Scholar]
- ELBASHIR S.M., LENDECKEL W., and TUSCHL T. (2001b). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONG W., REN Y., XU Q., WANG Y., LIN D., ZHOU H., and LI T. (2006). Integrated siRNA design based on surveying of features associated with high RNAi effectiveness. BMC Bioinformatics 7, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONG W., REN Y., ZHOU H., WANG Y., KANG S., and LI T. (2008). siDRM: an effective and generally applicable online siRNA design tool. Bioinformatics 24, 2405–2406 [DOI] [PubMed] [Google Scholar]
- GOSSEN M., and BUJARD H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLEN T., AMARZGUIOUI M., WIIGER M.T., BABAIE E., and PRYDZ H. (2002). Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 30, 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU X., HIPOLITO S., LYNN R., ABRAHAM V., RAMOS S., and WONG-STAAL F. (2004). Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res. 32, 4609–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHVOROVA A., REYNOLDS A., and JAYASENA S.D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 [DOI] [PubMed] [Google Scholar]
- KRETSCHMER-KAZEMI FAR R., and SCZAKIEL G. (2003). The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 31, 4417–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRUEGER U., BERGAUER T., KAUFMANN B., WOLTER I., PILK S., HEIDER-FABIAN M., KIRCH S., ARTZ-OPPITZ C., ISSELHORST M., and KONRAD J. (2007). Insights into effective RNAi gained from large-scale siRNA validation screening. Oligonucleotides 17, 237–250 [DOI] [PubMed] [Google Scholar]
- LARSSON E., SANDER C., and MARKS D. (2010). mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol. 6, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU Z.J., and MATHEWS D.H. (2008). OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 36, W104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATVEEVA O., NECHIPURENKO Y., ROSSI L., MOORE B., SAETROM P., OGURTSOV A.Y., ATKINS J.F., and SHABALINA S.A. (2007). Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 35, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAITO Y., and UI-TEI K. (2012). siRNA Design Software for a Target Gene-Specific RNA Interference. Front. Genet. 3, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OVCHARENKO D., JARVIS R., HUNICKE-SMITH S., KELNAR K., and BROWN D. (2005). High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA 11, 985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OVERHOFF M., ALKEN M., FAR R.K., LEMAITRE M., LEBLEU B., SCZAKIEL G., and ROBBINS I. (2005). Local RNA target structure influences siRNA efficacy: a systematic global analysis. J. Mol. Biol. 348, 871–881 [DOI] [PubMed] [Google Scholar]
- REN Y., GONG W., ZHOU H., WANG Y., and XIAO F., LI T. (2009). siRecords: a database of mammalian RNAi experiments and efficacies. Nucleic Acids Res. 37, D146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- REYNOLDS A., LEAKE D., BOESE Q., SCARINGE S., MARSHALL W.S., and KHVOROVA A. (2004). Rational siRNA design for RNA interference. Nat. Biotechnol 22, 326–330 [DOI] [PubMed] [Google Scholar]
- ROSENBLOOM K.R., DRESZER T.R., LONG J.C., MALLADI V.S., SLOAN C.A., RANEY B.J., CLINE M.S., KAROLCHIK D., BARBER G.P., CLAWSON H., et al. (2012). ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 40, D912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIEL G., and CIBELLI G. (2002). Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell Physiol. 193, 287–292 [DOI] [PubMed] [Google Scholar]
- TRUSS M., SWAT M., KIELBASA S.M., SCHAFER R., HERZEL H., and HAGEMEIER C. (2005). HuSiDa–the human siRNA database: an open-access database for published functional siRNA sequences and technical details of efficient transfer into recipient cells. Nucleic Acids Res. 33, D108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UI-TEI K., NAITO Y., TAKAHASHI F., HARAGUCHI T., OHKI-HAMAZAKI H., JUNI A., UEDA R., and SAIGO K. (2004). Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32, 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.