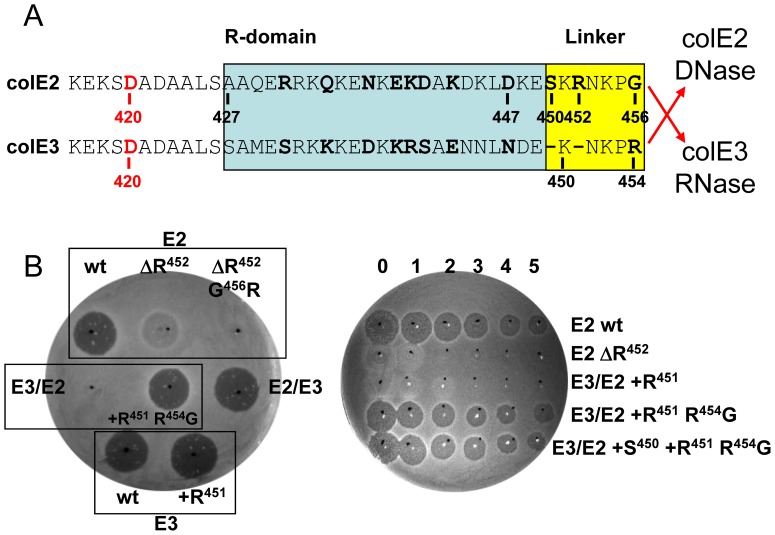
Figure 4. Toxicity of wild-type, mutant and hybrid-colicins E3/E2 and E2/E3 derivatives.
(A) Alignment of the C-terminal ends of the R domain (blue box) and the linker regions (yellow box) [15], [16] of colE2 and E3. Amino acids that are specific for either DNase or RNase E-type colicins are shown in bold. Hybrid E2 and E3 colicins were created by in-frame fusion between the upstream R domains and linkers and the downstream nuclease domains as indicated by red arrows. FtsH-dependent processing sites in colicins E2 and E3 at or close to residue D420 (amino acid in red) are shown. (B) Comparison of the cytotoxicity of wild-type (wt; E2 or E3), mutant and hybrid colicins (E3/E2 or E2/E3) by growth inhibition test. Mutant derivatives of the wild-type or hybrids are grouped with their parental strain (black rectangles). R452 was deleted in E2 (ΔR) and was combined with the G456R substitution. In E3/E2 hybrid an R added at position 451 (+R) was combined with the substitution R454G and an S added at position 450 (+S). The in vivo assay when necessary was quantified by spotting aliquots of undiluted colicin (numbered 0) and serial 10-fold dilutions up to 10−5 (numbered 1 to 5) directly onto a lawn of OmpT-deficient AD202 strain (right panel). Dark halos indicate a clear zone of growth inhibition (toxicity), and no clearing indicates resistance to colicin. Turbid zones indicate marginal toxicity.