Figure 1.
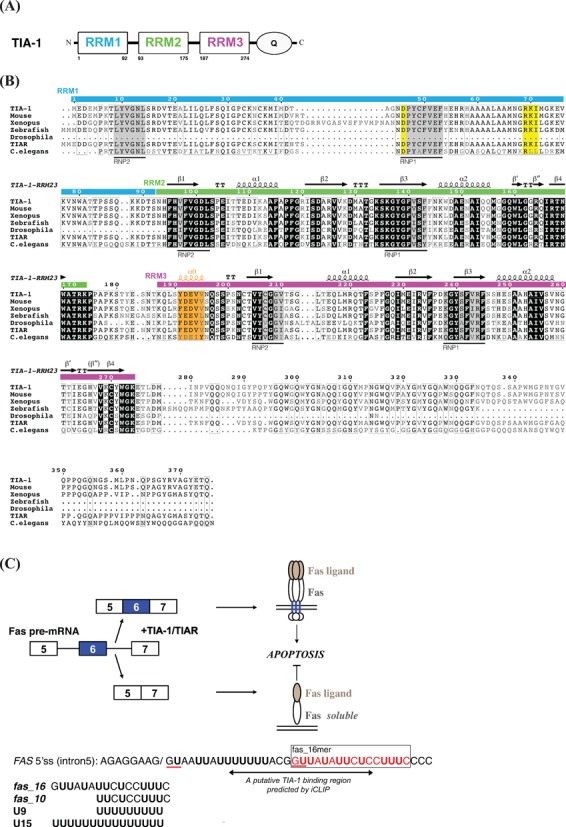
(A) Schematic representation of human TIA-1 domains. (B) Sequence alignment of TIA-1 and TIAR from different species. Original sequences used in the analysis: residues 1–375 of Homo sapiens TIA-1, residues 1–377 of Mus musculus TIA-1, residues 41–427 of Xenopus laevis TIA-1, residues 1–342 of Danio rerio (zebrafish) TIA-1, residues 166–356 of Drosophila melanogaster TIA-1, residues 1–375 of Homo sapiens TIAR and residues 45–373 of Caenorhabditis elegans TIAR are compared and aligned by ClustalW2 (21) and plotted by ESPript 2.2 (22,23). The secondary structure indicated corresponds to the solution structure of human TIA-1-RRM2,3. The RNP1 and RNP2 motifs are highlighted with gray background, the negatively charged residue in RNP1 and the RXI/V/L motif of RRM1 indicative of a potential UHM domain are shown with yellow background. The residues in RRM3 helix α0 are colored orange. (C) Alternative splicing of human FAS exon 6. The putative TIA-1 binding region in FAS intron 5 indicated by a double-arrow is predicted by iCLIP analyses, i.e. 10–28 nucleotides downstream of exon/intron boundaries. The GU motifs corresponding to 5′ splice sites in FAS intron 5 RNA are underlined in red. RNA sequences of the 16-mer pre-RNA ligand in FAS intron5 (fas_16) and a uridine-rich RNA (U9), used in the present study are indicated.
