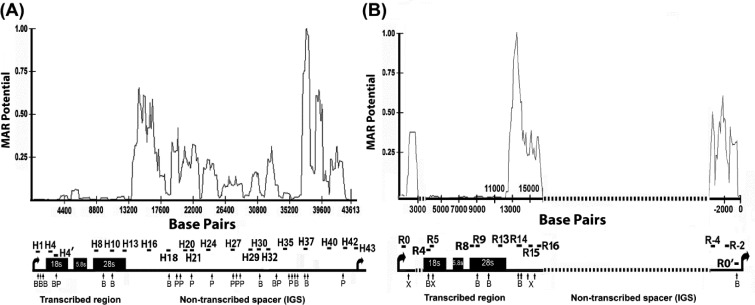
Figure 1.
The non-transcribed intergenic spacer (IGS) region of rRNA genes has an enhanced propensity for nuclear matrix attachment. The line graphs show MAR potential score in relation to nucleotide position for the rDNA repeat sequence from human (A) and rat (B). The lower panels in each figure part show schematic depictions of rRNA gene repeats. Vertical arrows below the rDNA repeat represent the locations of cleavage sites for BamHI (B) and PstI (P) in human (part A) and for BamHI (B) and XhoI (X) in rat (part B). The locations of amplified rDNA regions are shown by horizontal bars. The sequences of primer sets used for amplification of human and rat rDNA are shown in Supplementary Tables S1 and S2. The dotted line in rat rRNA gene represents sequences that are not available from GenBank. Human rDNA sequence, derived from GenBank number U13369, and (B) rat rDNA sequence, derived from accession numbers X04084, X03838, X61110, X00677, X16321, V01270 and X03695, were used. Matrix attachment potential was predicted by the MAR-Wiz software (57) as implemented at http://genomecluster.secs.oakland.edu using a window size of 1000 bp stepped at 100-bp intervals. Scores in excess of 0.6 are generally considered as strong indicators of nuclear matrix binding potential.