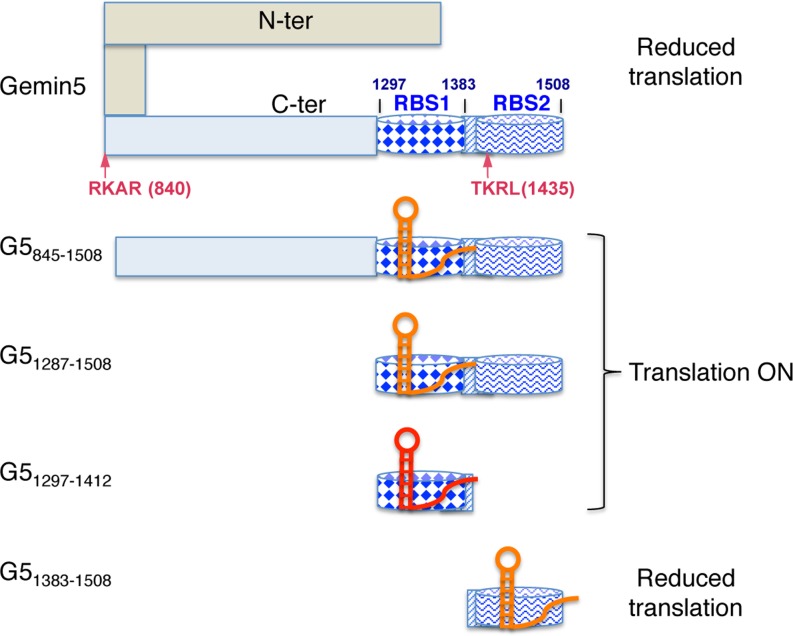
Figure 8.
Hypothesized model for the differential effect of Gemin5 truncated polypeptides on translation control. The C-terminal region of Gemin5 starting at residue 845 is shown in pale blue. Within this region, dark blue barrels depict the RNA-binding sites RBS1 (diamonds) and RBS2 (wavy line), respectively. The striped rectangle located between these sites depicts the overlapping region of the polypeptides analysed in this study, which was partially deleted in construct G5Δ1365–1394. Arrows depict the RKAR (840) and TKRL (1435) recognition sites of the FMDV L protease. Domain 5 of the FMDV IRES is shown as a red or orange hairpin, indicating the higher or lower efficiency of binding to the Gemin5 polypeptides based on gel-shift or UV-crosslinking assays. In this model, we hypothesize that the IRES RNA binds to RBS2 on the most C-terminal fragment G51383–1508 and the full-length Gemin5 protein reducing translation efficiency, while binding of the IRES RNA to RBS1 in proteins G5845–1508, G51287–1508 and G51297–1412 does not adversely affect internal initiation of translation. Details are provided in the text.