Figure 5.
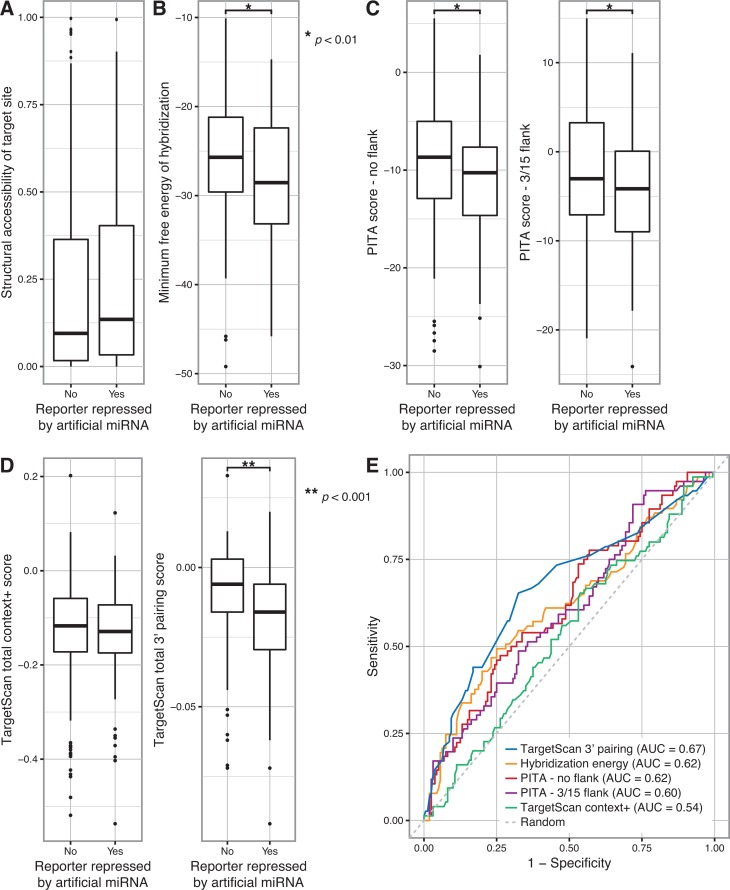
Artificial miRNA activity is modestly predicted by base pairing at the miRNA 3′ end. (A) The probability of each artificial miRNA seed match site in the PC and GLS 3′ UTR reporter genes being structurally accessible (i.e. single-stranded) was calculated, and the most accessible site was determined for each miRNA/reporter pair. Boxplots represent the structural accessibility probability values for miRNA/reporter pairs in which the miRNA did or did not repress the reporter. A t-test yielded P > 0.05. (B) The minimum free energy of hybridisation between each artificial miRNA and the PC and GLS 3′ UTRs was calculated, and the hybridisation site with the lowest free energy was determined for each miRNA/gene pair. Boxplots represent the minimum free energy for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. A t-test with P < 0.01 is indicated by the single asterisk. (C) Target prediction scores were calculated using the PITA algorithm for each artificial miRNA and the PC and GLS 3′ UTRs, using settings that excluded flanking nucleotides (left) or considered 3 upstream and 15 downstream flanking nucleotides (right). Boxplots represent the scores for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. t-tests with P < 0.01 are indicated by a single asterisk. (D) TargetScan total context+ scores (left) were calculated for each artificial miRNA and the PC and GLS 3′ UTRs. The total sub-score for 3′ end base pairing, calculated as part of the context+ score, was analysed separately (right). Boxplots represent the indicated scores for miRNA/gene pairs in which the miRNA did or did not repress the corresponding 3′ UTR reporter. A t-test of the 3′ pairing score with P < 0.001 is indicated by the double asterisk. A t-test of the context+ score yielded P > 0.05. (E) The ability of the indicated measures of miRNA targeting to predict repression of PC and GLS 3′ UTR reporter genes by each artificial miRNA were compared by plotting sensitivity versus specificity. The area under the curve (AUC) for each prediction method is indicated. A random prediction is shown as a dashed line.