Figure 6.
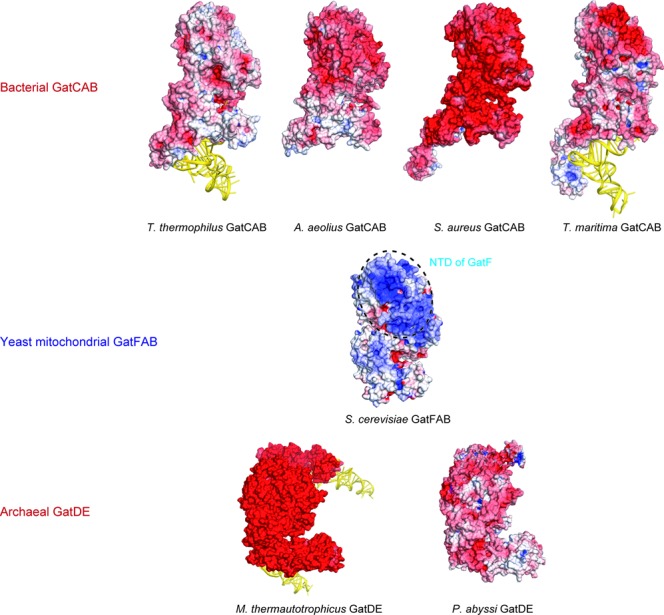
Electrostatic surface potential models of tRNA-dependent amidotransferases. Bacterial GatCABs (PDB IDs: 3KFU for T. thermophilus GatCAB-tRNAAsn complex, 3H0R for A. aeolius GatCAB, 3IP4 for S. aureus GatCAB and 3AL0 for T. maritima GatCAB-tRNAGln complex) are shown on the upper lane. S. cerevisiae GatFAB (this study) is shown on the center of the figure. The NTD of GatF is indicated by the dashed circle. Archaeal GatDEs (PDB IDs: 2D6F for M. thermautotrophicus GatDE-tRNAGln complex and 1ZQ1 for P. abyssi GatDE) are shown on the lower lane. Electrostatic surface potential is calculated by the program APBS (36).
