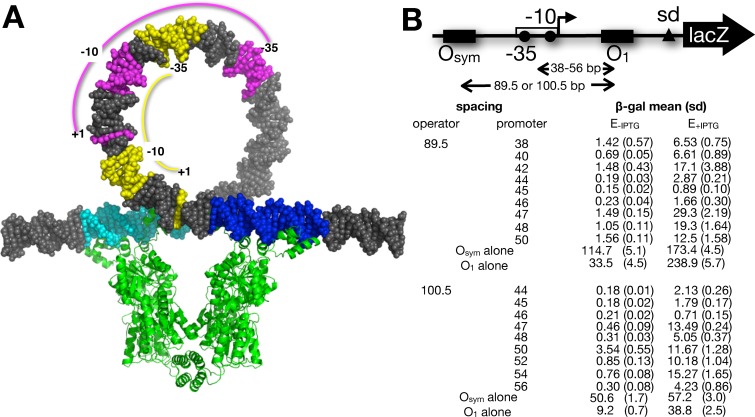
Figure 2.
DNA looping constructs and data. (A) Scale space-filling models showing lac loop geometry studied here. 100.5 bp DNA loop is shown stabilized by the lac repressor tetramer (green; pdb code 1Z04) engaged with upstream Osym (cyan) and downstream O1 (blue) operators in the conventional model. This conventional model depicts the simplifying assumption that lac repressor does not deform its bound operators, and depiction of this conventional model is not meant to imply that there are not superior loop models. The positions of the wild-type lac promoter −35, −10 and +1 elements (yellow) are contrasted with an example of promoter elements in this study (magenta) where promoter and operator do not overlap. The diagram was rendered using the 3D-DART tool (57). (B) β-galactosidase expression data from constructs with the indicated operator and promoter spacings (mean, standard deviation in parentheses). Promoter spacing indicates distance (bp) between the center of O1 and the center of the UV5 promoter. Operator spacing indicates distance (bp) between the centers of O1 and Osym operators. Results for constructs with single operators are shown.