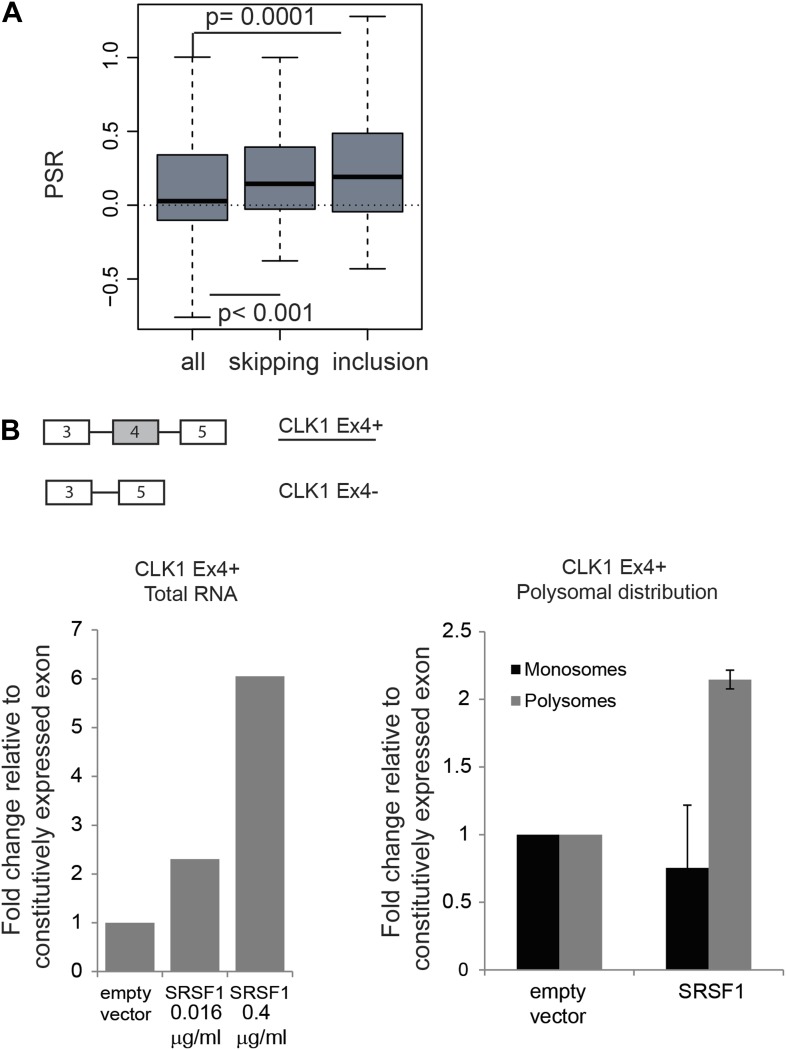
Figure 4. Coupling of alternative splicing and translational regulation.
(A) Correlation between SRSF1-induced changes in alternative splicing with polysomal distribution of those isoforms. Changes in alternative splicing induced by SRSF1 overexpression were determined by an exon-junction array. PSR: polysome shift ratio. (B) RT-qPCR analysis of the effect of SRSF1 on CLK1 alternative splicing and preferential polysomal association. The exon-intron structure of both isoforms is indicated (not to scale) and the CLK1 isoform that is an SRSF1 direct translational target is underlined. SRSF1-induced changes in CLK1 alternative splicing were determined and normalized to exon 7 (constitutive exon) levels (left panel). Two different concentrations of pCGT7-SRSF1 plasmid were used. Polysomal distribution of CLK1 mRNA isoforms upon SRSF1 overexpression normalized to exon 7 (constitutive exon) levels (right panel).