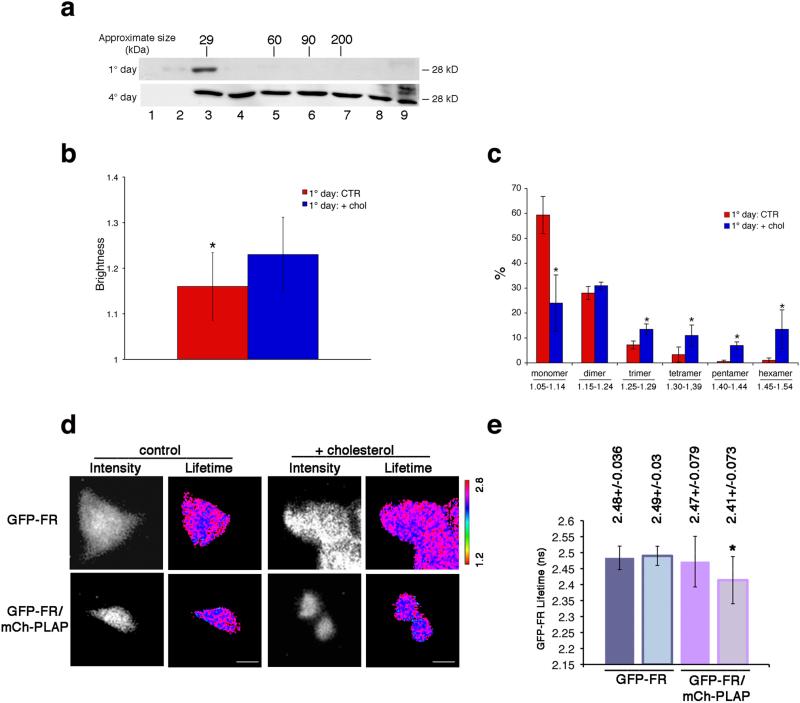
Figure 4. Cholesterol addition is necessary and sufficient to induce homo- and hetero-cluster of GPI-APs in non-polarized MDCK cells.
(a) MDCK cells expressing GFP-FR grown for 4 days (polarized) or 1 day (non-polarized) were treated with trypsin (to remove surface proteins) and then subjected to temperature block (at 19.5°C in the Golgi) in order to analyze exclusively the Golgi pool of proteins. Cell lysates were run on velocity gradient (see Methods). Fractions were collected from top (Fraction 1) to bottom (Fraction 9), TCA-precipitated, run on SDS-PAGE and GFP-FR was revealed by western blot. Molecular weight markers are indicated on top of the panels. (b-e) 1 day MDCK cells (control or upon cholesterol addition) were used for N&B and FLIM analysis. (b) Quantification of the brightness of GFP-FR from 3 independent experiments is plotted. Error bars, ± SD. *, p<0,0001. (c) Graphical representation of the percentage of pixels falling in the different classes of B values (from monomer to hexamer; see Supplementary Fig. 4g). Values are the mean of 3 independent experiments. Error bars, ± SD. *, p<0.05. (d) Intensity and mean fluorescence lifetime maps of GFP-FR alone or in combination with mCherry-PLAP. The lifetime scale is from 1.2 ns to 2.8 ns. Bars, 9 μm. (e) Histograms of GFP-FR lifetime alone (blue bars) or in combination with mCherry-PLAP (pink bars) in control conditions (colored bars) or upon cholesterol addition (pale colored bars). Experiments were performed 3×, n>15 cells. The error bars are the mean ± SD. *, p<0,005.