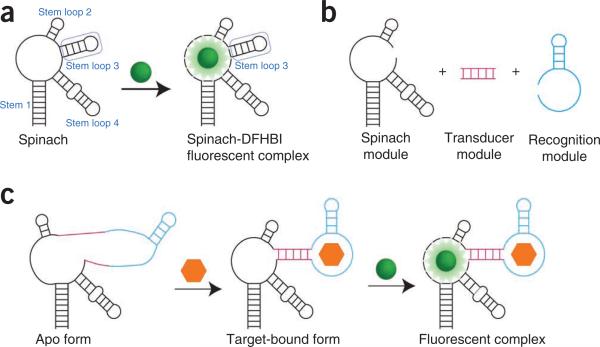
Figure 1.
Modular strategy for generating Spinach-based sensors. (a) Spinach is an RNA aptamer that binds a small-molecule dye called DFHBI (green ball). Both DFHBI and Spinach are nonfluorescent until binding occurs and activates the fluorescence of the Spinach-DFHBI complex. Stem loop 3 of Spinach can tolerate insertion of additional sequences, and it is the region that is modified to generate sensors. (b) In Spinach-based sensors, Spinach is modified to include a transducer region (magenta) and a recognition module (cyan). Recognition molecules are typically aptamers generated against a target ligand by SELEX2,3, but they can also be composed of riboswitch regions10,11 and naturally occurring RNAs3. Transducers of varied length and composition can be generated in order to optimize sensor function. (c) In the absence of DFHBI and ligand (orange hexagon), the Spinach-based sensor displays minimal fluorescence. However, upon target binding, the recognition module of the sensor folds and induces folding of the Spinach portion of the sensor. The Spinach-based sensor is then able to bind DFHBI and activate fluorescence.