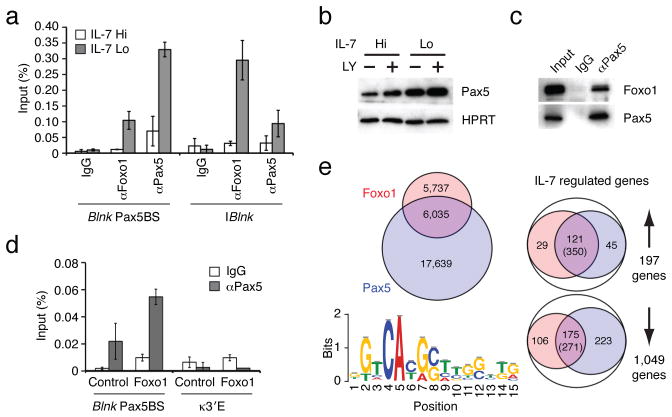
Figure 4. Foxo1 and Pax5 cooperate to induce Blnk expression upon attenuation of IL-7 signaling.
(a) ChIP analysis of Foxo1 and Pax5 binding to the denoted regulatory elements in the Blnk locus. ChIP was performed as in Fig. 1c. (b) Immunoblot analysis of Pax5. Cells were treated with vehicle or 53μg/mL of PI3K inhibitor LY294002 (LY). Protein expression was normalized with HPRT. (c) Immunoprecipitation of Pax5 complexes. Whole cell extracts were prepared from IRF-4,8−/− pre-B cells cultured under IL-7 Lo conditions for 48 h. Immunoprecipitation was performed using either control IgG or αPax5 antibodies, followed by immunoblot analysis with αFoxo1 or αPax5 antibodies. 5% of pre-immunoprecipitated samples were used for input. Data in a–c are represetative of three independent experiments. (d) Pax5 binding to the regulatory element in the Blnk locus in IRF-4,8−/− pre-B cells after transduction with Foxo1 retroviral vector as described above. ChIP assay was performed using control IgG or αPax5 antibodies. The error bars correspond to the standard deviation from the mean for three independent experiments. The κ3′E region is used as a negative control. (e) Comparative analysis of Foxo1 and Pax5 cistromes in IRF-4,8−/− pre-B cells. ChIPseq analysis of Pax5 binding was performed as in Fig. 2a. Left; A venn diagram comparing Foxo1 cistrome (red) with the Pax5 cistrome (blue). Based on 17,639 Pax5 binding peaks, de novo motif analysis was performed with the top 500 peaks as in Fig. 2a. Sequence logo depicts the Pax5 motif (P-value for motif significance; P = 6.3E-05) that was present in 341 of the 500 sequences analyzed. Right; Union analysis comparing the Foxo1 and Pax5 cistromes with IL-7 regulated genes as described in Fig. 2b. The number of genes in each category is shown. Numbers in parentheses indicate DNA sequences that display coincident binding of Foxo1 and Pax5 in IL-7 regulated genes that are co-targeted by both transcription factors.