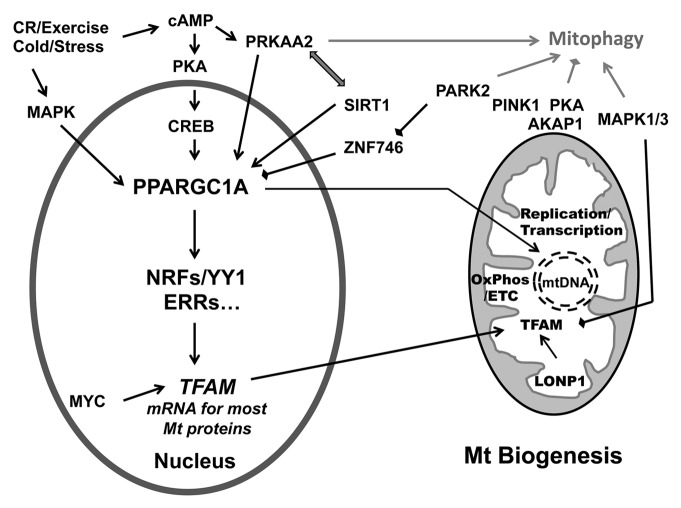
Figure 2. The regulation of mitochondrial biogenesis by the PPARGC1A-NRF1-TFAM pathway. PPARGCA is activated in response to environmental (cold), physiological [exercise or caloric restriction (CR)] or pathological stimuli (oxidative stress). One major pathway involves PKA phosphorylation of CREB that in turn activates PPARGC1A and multiple transcription factors (NRFs, YY1, ERRs, etc.) to result in the synthesis of both nDNA-encoded mitochondrial proteins and mitochondrial transcriptional regulators such as TFAM. TFAM in turn functions to stabilize mtDNA and promote the synthesis of mtDNA-encoded subunits of the electron transport chain (OxPhos/ETC). cAMP-associated phospho-activation and/or deacetylation by SIRT1 increases the activity of PPARGC1A. PPARGC1A can also traffic to mitochondria, where it interacts with TFAM at the mtDNA D-loop. Several major signaling pathways such as MAPK1/3, MYC, MAPK12, and PARK2 are also implicated in modulating mitochondrial biogenesis at different levels. In addition, the PRKAA2, PINK1-PARK2, PKA, and MAPK1/3 pathways also regulate mitophagy, potentially serving to coordinate overall changes in mitochondrial content.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.