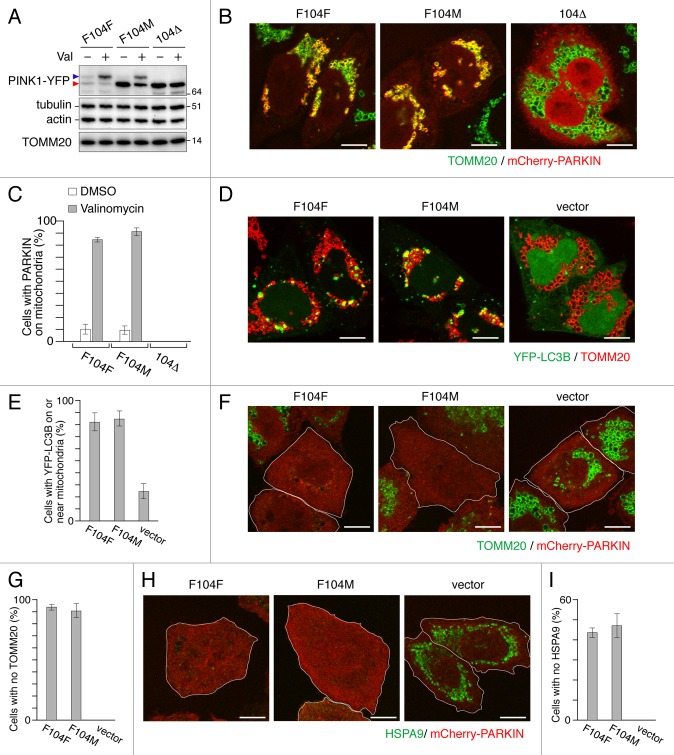
Figure 4. PINK1F104M mutant has the ability to recruit PARKIN to depolarized mitochondria followed by mitophagy. (A) Immunoblotting of HeLa cells expressing PINK1-YFP variants (F104F, F104M and 104Δ) treated with or without valinomycin (Val) for 3 h. Blue and red arrowheads represent the full-length and the cleaved forms of PINK1-YFP, respectively. (B and C) pink1−/− MEF cells were cotransfected with PINK1-YFP variants and mCherry-PARKIN. After 12 h of transfection, cells were treated with DMSO or valinomycin for 4 h. (B) Cells were then immunostained with anti-TOMM20. Scale bars: 10 μm. (C) Cells with mCherry-PARKIN on mitochondria were quantified. Error bars were calculated from three independent experiments. (D and E) pink1 KO HeLa cells stably expressing mCherry-PARKIN were cotransfected with YFP-LC3B and untagged either PINK1 WT(F104F) or PINK1(F104M) or pcDNA3.1(+) (vector). Cells were treated with valinomycin for 3 h, and immunostained with anti-TOMM20. (D) Scale bars: 10 μm. (E) Cells with YFP-LC3B on or near mitochondria were quantified. Error bars represent the results from three independent experiments. (F–I) pink1 KO HeLa cells stably expressing mCherry-PARKIN were transfected with WT PINK1-YFP (F104F), PINK1F104M-YFP or pEYFP-C1 (vector). Cells were treated with valinomycin for 12 h, and then immunostained with anti-TOMM20 or anti-HSPA9 antibodies. (F and H) YFP-positive cells are shown in white outlines. Scale bars: 10 μm. (G and I) Cells with complete degradation of TOMM20 or HSPA9 were quantified. Error bars represent the results from three independent experiments.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.