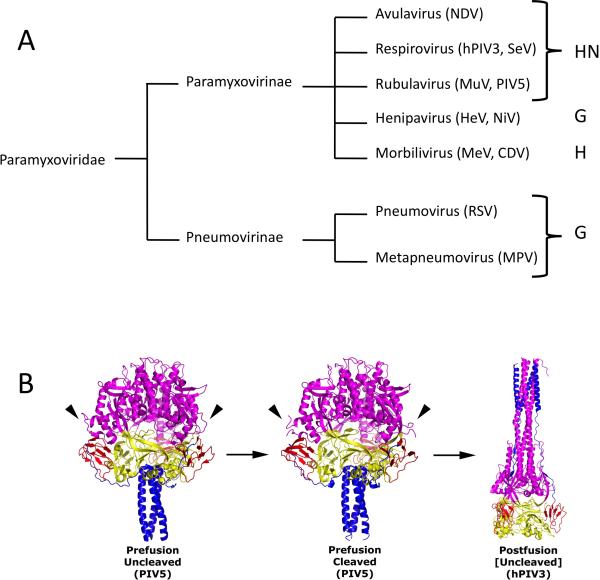
Figure 1. The attachment and fusion glycoproteins of the Paramyxoviridae.
(A) Pseudo phylogenetic tree of examples of the Paramyxoviridae. No evolutionary distance is implied by the lines. Genera are listed to the right and associated attachment glycoprotein abbreviations are indicated to the far right. NDV, Newcastle disease virus; HeV, Hendra virus; NiV, Nipah virus; MeV, measles virus; CDV, canine distemper virus; MuV, mumps virus; PIV5, parainfluenza virus 5; SeV, Sendai virus; PIV3, parainfluenza virus 3; RSV, respiratory syncytial virus; MPV, metapneumovirus. (B) Structure of distinct conformational states of the F protein ectodomain. Side-view ribbon diagrams of prefusion PIV5 F protein in its uncleaved (PDB ID code: 2B9B) [67] and protease-cleaved states (PDB ID code 4GIP) [7], as well as the uncleaved postfusion structure of the hPIV3 F ectodomain (PDB ID code: 1ZTM) [68]. Arrowheads indicate the position of protease cleavage sites in the prefusion structures for two of the three chains (the third is hidden from view). Arrows indicate the progression of F through its various conformational states. An exception for the uncleaved postfusion hPIV3 F structure is noted by square brackets as cleavage activation is a biological requirement for fusion activity, although not for refolding of the F ectodomain into the postfusion form. Each structure is color-coded by domain (DI, yellow; DII, red; DIII, magenta; HRB, blue). Reproduced from Welch and coworkers (2012) [7].