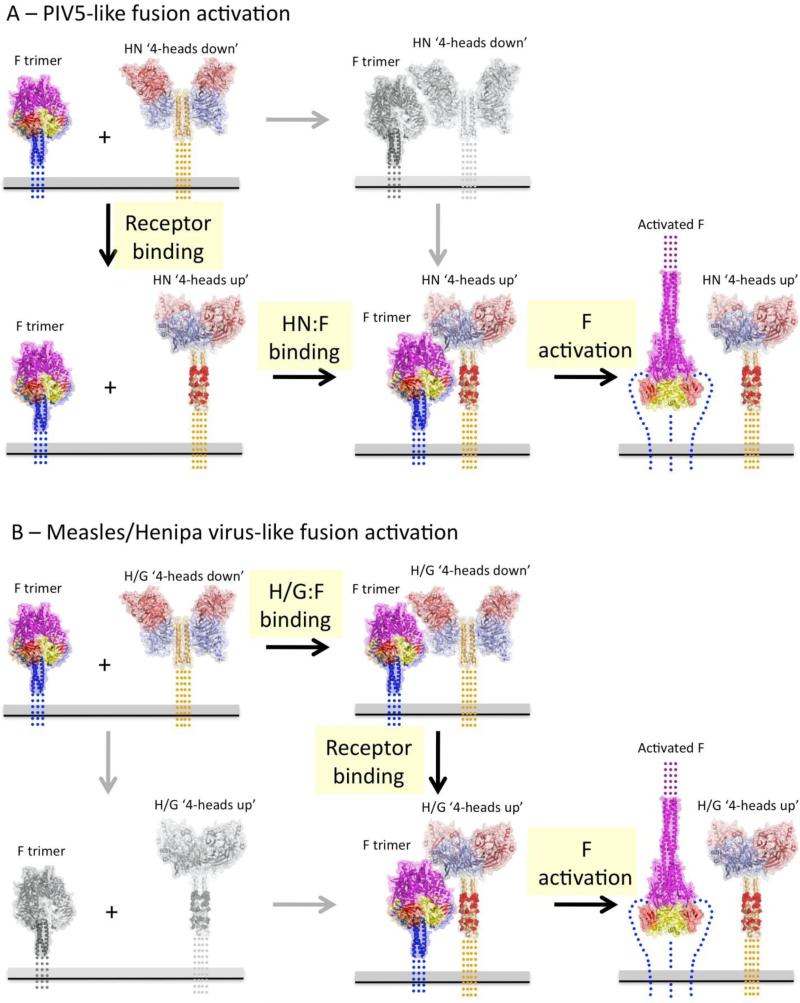
Figure 5. Models for receptor-dependent fusion activation for paramyxoviruses within the Paramyxovirinae subfamily.
Current experiments indicate that the attachment glycoproteins from many paramyxoviruses within the Paramyxovirinae subfamily adhere to a general set of features. The attachment glycoproteins are structurally related and have a globular, C-terminal RBD that can interact with various receptors with diverse complex architectures. The attachment glycoprotein specificity for F activation lies within the N-terminal, tetrameric stalk and the ‘headless’ stalk domains constitutively activate F. The F proteins fold to a functional, prefusion state in the absence of the attachment proteins, indicating that a stabilizing clamp is not required for their function. The structures of two different attachment glycoprotein ectodomains show a stalk-RBD interaction that occludes residues implicated in F binding and activation. Stalk residues that are separately responsible for F protein binding and F protein activation have been identified, suggesting two distinct steps in F activation, which could be energetically different in different viruses. These data indicate that F binding is not absolutely required for F stabilization, disfavoring a clamp model, and that F binding Is necessary but insufficient for F activation. The data appear most consistent with a provocateur model, where residues in the HN/H/G stalk domain are specifically involved in destabilizing the prefusion F protein to undergo conformational changes that drive membrane fusion. Two related models that capture these features for PIV5-like and Henipa/measles virus-like activation pathways are shown in panels (A) and (B). The grey panels and arrows indicate low probability pathway steps based on available functional data. Red atoms in the post-receptor bound HN/H/G stalk represent F interacting and activating residues. ‘Activated” prefusion F is indicated by CPK atoms for residues that undergo large conformational changes to the post-fusion conformation (far right panels). (A) PIV5-like fusion activation involves weak or no interaction between prefusion F and prefusion HN proteins. Binding to receptors on target cells would fully expose the stalk region, allow F to bind and then to be activated at the appropriate time and place. (B) Henipa/measles virus-like fusion activation involves preformed complexes between prefusion F and H/G proteins that does not expose activating residues with the H/G stalk to F. Receptor binding would reorient the H/G RBDs, allowing full binding to the stalk and subsequent destabilization of prefusion F. The two pathways show significant functional conservation, as revealed by extensive comparative data that has been collected. Differences in the nature and stability of the prefusion HN/H/G complexes with F may reflect evolutionary differences in the strength of these interactions but these differences are not in and of themselves indicative of fundamentally different activation mechanisms or pathways. On the contrary, the accumulating data suggest an overall conserved mechanism of receptor-dependent activation across the Paramyxovirinae subfamily of the paramyxoviruses.