Figure 4.
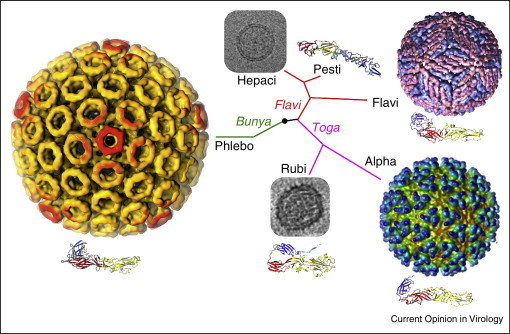
Structural relationships of viruses that contain class II fusion proteins. The class II fold is highly conserved in flaviviruses, alphaviruses and phleboviruses, even though these viruses differ in their genomic organization, coding strategies and outer protein shell assemblies. These three genera have in common that they have lifecycles that alternate between vertebrate and arthropod hosts. Rubella virus (RV) E1 has the most divergent class II fold even though rubella belongs to the same family as alphaviruses (Togaviridae). Glycoprotein E2 from the pestivirus bovine viral diarrhea virus has a novel fold even though pestiviruses belong to the same family as flaviviruses (Flaviviridae) [9••, 10••]. Rubella virus and pestiviruses, and their close relatives the hepaciviruses, have in common that they infect strictly vertebrate hosts, and also that they do not form rigid icosahedral outer protein shells. Thus, structural conservation in viral fusion proteins does not correlate with overall phylogenetic relatedness. The virus particles shown here are, clockwise from top right, dengue virus, Semliki Forest virus, RV, Rift Valley fever virus and hepatitis C virus (HCV). The electron micrographs of RV [55•] and HCV [61] are not drawn to scale with the particles in color. The phylogenetic tree is based on qualitative structural and genetic relationships between envelope proteins and is not based on a quantitative phylogenetic analysis.
