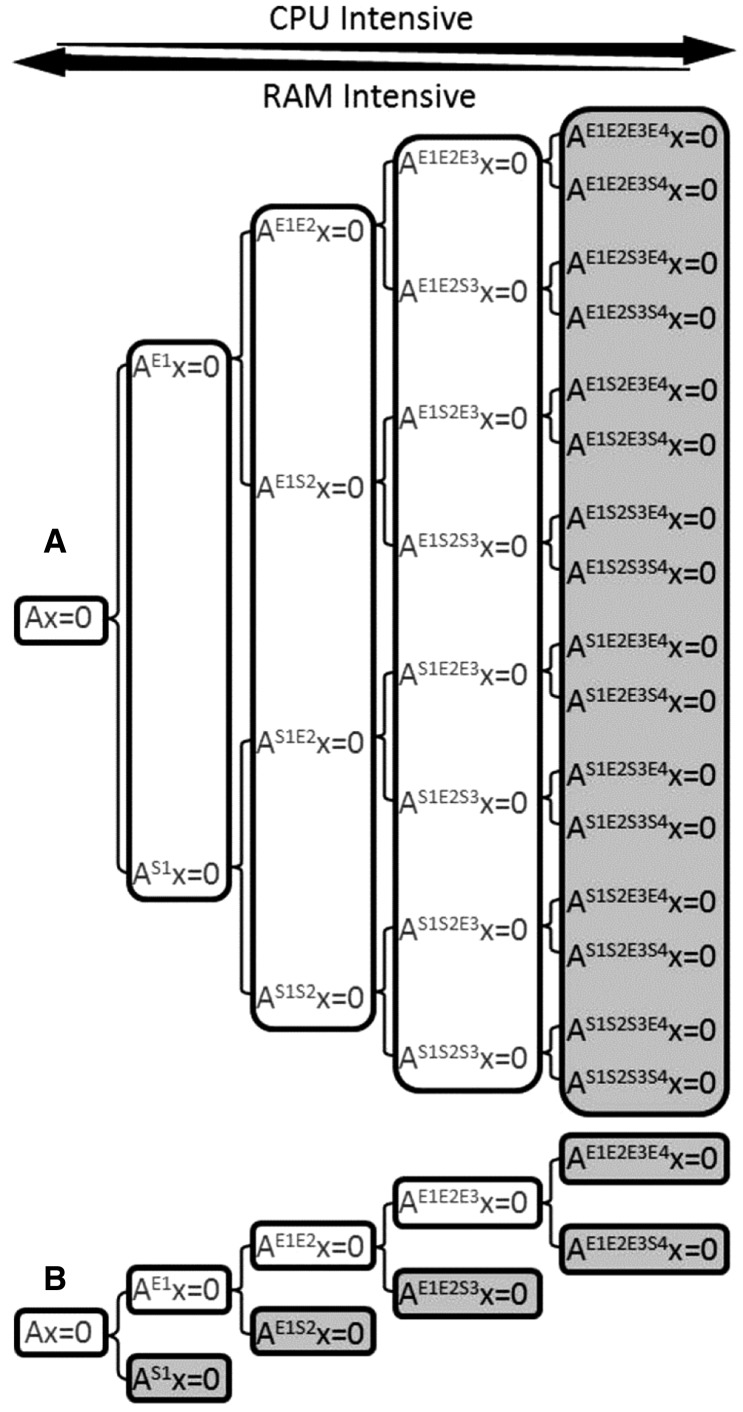
Fig. 5.
Comparison of two network splitting approaches. The basic splitting approach (A) uses combinations of enforced/suppressed reactions to create calculable subnetworks where the number of subnetworks increases exponentially with the number of reactions for splitting (Klamt et al., 2005). The iterative splitting approach (B) only generates subnetworks when the previous ones are intractable (white) until all EFMs for all subnetworks in the branch can be enumerated (gray) (Klamt et al., 2005). Both approaches shift the burden of computation from a RAM-limited problem to a CPU-limited problem as the subnetworks become smaller but greater in number. The example uses R = 4 reactions for splitting with each iteration considering one additional reaction (i.e. r = 1) for a total of i = 4 iterations. Superscripts refer to the enforced (E) or suppressed (S) reactions in the labeled subnetworks