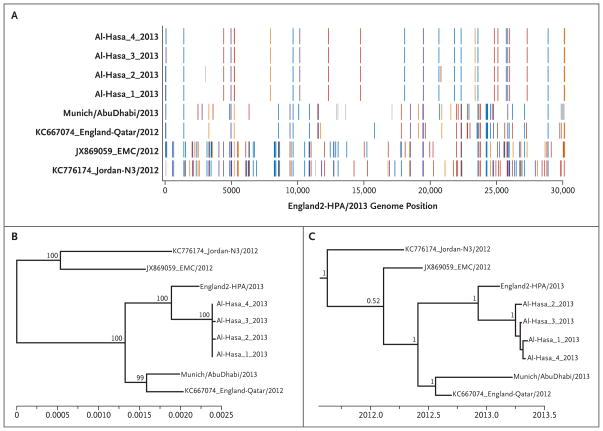
Figure 4. Phylogenetic Analysis of the Sequences of All Genes Identified in Four Patients Infected with MERS-CoV.
Panel A shows single-nucleotide differences (vertical colored bars) between the England2 genome and the four Al-Hasa genomes as well as the four additional full genomes available; gray indicates a gap in the query sequence, orange a change to A, crimson a change to T, blue a change to G, and purple a change to C. The reference genomes we used were from a Jordanian patient in April 2012 (Gen-Bank accession number, KC776174), EMC/2012 from a Saudi Arabian patient in July 2012 (JX869059),19 England/Qatar/2012 from a London Qatari patient in September 2012 (KC667074),22 England2 from a patient who had traveled to Pakistan and Saudi Arabia in February 2013,13 and the Munich/AbuDhabi sequence from a patient from the United Arab Emirates in March 2013.23 Panel B shows an unrooted maximum-likelihood phylogeny inferred under a generalized-time-reversal (GTR)+Gamma substitution model that compares the five previously identified Middle East respiratory syndrome (MERS) genomes with the four Al-Hasa genomes. Bootstrap values are shown for the highly supported nodes. Panel C shows a time-resolved maximum clade credibility tree for the five previously identified genomes and the four Al-Hasa MERS coronavirus genomes. Posterior probability values are shown for nodes with posterior support greater than 0.5. Findings are consistent with previously published estimates.24