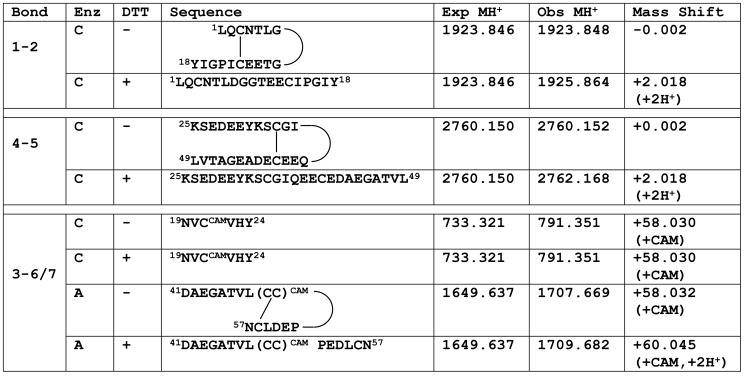
Figure 1. Summary of mass spectral analysis in PMF-G disulfide bonding pattern determination.
Mass spectral analyses was performed on the three-disulfide species of PMF-G purified by RP-HPLC. Differential treatment included proteolytic enzyme (Enz; chymotrypsin [C] or AspN [A]), reduction with dithiothreitol (DTT), and alkylation with iodoacetamide (addition of a carboxyamidomethyl (CAM) group). Observed monoisotopic masses were compared to theoretical masses with no free sulfhydryls, and mass shifts used to determine peptide modification. All assignments were confirmed by analysis of the fragmented ion series.