Abstract
CITED2 was identified as a cardiac transcription factor which is essential to the heart development. Cited2-deficient mice showed cardiac malformations, adrenal agenesis and neural crest defects. To explore the potential impact of mutations in CITED2 on congenital heart disease (CHD) in humans, we screened the coding region of CITED2 in a total of 700 Chinese people with congenital heart disease and 250 healthy individuals as controls. We found five potential disease-causing mutations, p.P140S, p.S183L, p.S196G, p.Ser161delAGC and p. Ser192_Gly193delAGCGGC. Two mammalian two-hybrid assays showed that the last four mutations significantly affected the interaction between p300CH1 and CITED2 or HIF1A. Further studies showed that four CITED2 mutations recovered the promoter activity of VEGF by decreasing its competitiveness with HIF1A for binding to p300CH1 and three mutations decreased the consociation of TFAP2C and CITED2 in the transactivation of PITX2C. Both VEGF and PITX2C play very important roles in cardiac development. In conclusion, we demonstrated that CITED2 has a potential causative impact on congenital heart disease.
Introduction
Congenital heart disease (CHD) is a most common defect caused by abnormal cardiac formation in fetuses and has become the leading reason of childhood mortality with an incidence around 1%[1]–[3]. In the past decades, a series of CHD-causing genes have been identified such as NKX2-5, TBX5, GATA4 and CITED2 [4]–[6]. It has been confirmed that their mutations can cause cardiac malformations through affecting the transcription activity of critical genes involved in heart development pathways.
CITED2 (Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2) is one member of a new conserved family of transcriptional activators which includes four members: CITED1 (Msg1), CITED2 (Mrg1/p35srj), CITED3 and CITED4 (Mrg2) [7]. CITED2 is a nuclear protein which binds closely to the CH1 region of p300 and CBP by its CR2 region (including a conserved 32-amino acid sequence [8]). Meanwhile, many other transcription factors and transcription regulating factors such as HIF1A, RXRα, NFk, Mdm2, Ets-1 and Stat2 also bind to the CH1 region of CBP/p300 [9], [10]. Thus CITED2 may act as a pivotal transcriptional modulator to regulate the expression of some specific genes. For example, CITED2 decreased the expression of HIF1A (Hypoxia Inducible Factor 1) through its competitive binding to CBP/p300CH1 [11], [12], consequently interfering the transcription of genes induced by HIF1A such as VEGF (vascular endothelial growth factor) [13]. It has been confirmed that the overexpression of vegf is the main factor resulting in cardiac malformation in cited2 -/- mice [14].
Besides being a transcriptional repressor of HIF1A, CITED2 acts as a transcriptional coactivator of TFAP2 (transcription factor AP2, also called Tcfap2) [15]. Mutations of TFAP2A and TFAP2B result in neural tube, cranial ganglia defects and cardiac malformations [16], [17]. This suggested that the coactivation of TFAP2 with p300, CITED2 and CREBBP is essential for the normal development of those structures. As a critical transcription factor, TFAP2 can affect the transcription of many genes, including PITX2C (Paired-Like Homeodomain 2 C)which is critical in Nodal-PITX2C pathways [18]. In addition, it has been detected that TFAP2 isoforms and CITED2 work together on the PITX2C promoter1 which controls the expression of PITX2C in the heart of embryonic mice. The mice experiments already indicated that knocking out pitx2c gene can lead to valve defects, body wall dysraphism, gastroschisis, ectopia cordis and other multiple organs polymorphous defects [19].
CITED2 gene mutation in human congenital heart disease was first reported by Sperling et al [20] in 2005. They identified 3 mutations which alter the amino acid sequence and studied their association with HIF1A and TFAP2C. Their study confirms that CITED2 is an important transcription factor in heart development and provides new insights into the molecular mechanism of congenital heart defects. Later, Yang et al found 3 new mutations in Chinese patients with congenital heart disease (2010) [21]and Chen et al [22]demonstrated another 3 new mutations in European CHD patients. Recently, Xu et al found 3 CITED2 gene mutations, their research showed that CITED2 gene mutations and methylation may play an important role in CHD. In their study, most of these mutations were in SRJ region. The mutations in our study were identified for the first time and located in SRJ region as well. Our work aimed to determine whether the new mutations also affect HIF1A or TFAP2C and finally lead to an abnormal expression of VEGF or PITX2C which play an important role in heart development.
Materials and Methods
Ethics statement
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the National Research Institute for Family Planning. Written informed consent was obtained from patients' parents or guardians.
Subjects
The study population comprised 700 patients who were diagnosed with CHD based on anthropometric measurement, physical examination for malformation and dysmorphism, and radiological evaluation. The patients with a phenotype of VSD, TOF and ASD accounted for 43.71%, 8.42% and 12% respectively. 250 unrelated healthy children were used as controls. Peripheral blood was collected from each affected individual and their parents and controls were from 6 months to 12 years old and most of them volunteered to participate in the study.
We sequenced the whole CITED2 ORF in 700 CHD patients (Table 1) and 250 healthy controls recruited from Lanzhou University, Beijing Children's Hospital, Zhengzhou Children's Hospital, Henan provincial Chest Hospital and Children's Hospital of Fudan University.
Table 1. Patients with congenital heart disease included in the study.
| Phenotype | Total(n = 700) |
| Ventricular septal defect(VSD) | 306 |
| Tetralogy of Fallot(TOF) | 59 |
| Atrial septal defect(ASD) | 84 |
| Patent ductus arteriosus(PDA) | 21 |
| Pulmonal atresia or stenosis(PS) | 21 |
| double outlet right ventricle(DORV) | 11 |
| Aortic coarctation(COA) | 4 |
| Pulmonary hypertension(PH) | 2 |
| Other complex cardiac malformations | 192 |
Mutational analysis and bioinformatics
Genomic DNA was extracted from peripheral blood leukocytes using standard methods. The human CITED2 gene is located on 6q24.1 and is encoded by two exons. One of the exons and splice sites of CITED2 were amplified by polymerase chain reaction (PCR) using two pairs of CITED2 gene-specific primers (Table 2). PCR products were sequenced using the appropriate PCR primers and the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and run on an automated sequencer, ABI 3730XL (Applied Biosystems), to perform mutational analysis.
Table 2. Primers used for PCR.
| Name | Primer pair |
| Primers for CITED2 | F CCGGCTGTGTTATGAGTGGTAG |
| R AGTTGGGGGTTTGATTTCTTTC | |
| Middle Primer for CITED2 | TCGGAAGTGCTGGTTTGTC |
| Primers for P140S | F TGCCGGATTTGCACTCTGCTGCA GGCCAC |
| R GTGGCCTGCAGCAGAGTGCAAAT CCGGCA | |
| Primers for S183L | F GCTCTGGCAGCAGCTTGGGCGGCG |
| R CGCCGCCCAAGCTGCTGCCAGAGC | |
| Primers for S196G | F AACAGCGGCGGCGGCGGCGGCAGCG GCAACA |
| R TGTTGCCGCTGCCGCCGCCGCCGCC GCTGTT | |
| Primers for Ser161delAGCAGC | F TGCAACCCCAAGCACGGCGGCAGCA GCACC TGCAACCCCAAGCACGGCGGCAGCAGCACC |
| R GGTGCTGCTGCCGCCGTGCTTGGGG TTGCA | |
| Primers for Ser192_Gly193delAGCGGC | F CGCGGGCAGCAGCAACGGCGGCAGC GGCAGCGGCAACAT |
| R ATGTTGCCGCTGCCGCTGCCGCCGTT GCTGCTGCCCGCG | |
| pEGFP-CIITED2 | F GGGGTACCATGGCAGACCATATGATG |
| R CGGGATCCCGACAGCTCACTCTGCTGG | |
| pCDNA3.1(+)-CITED2 | F CGGGGTACCTATGGCAGACCATATGA TGGC |
| R TGCTCTAGAGTCAACAGCTCACTCTGCTG | |
| pCMX-GAL4-CITED2 | F CGGATATCAATGGCAGACCATATGA TGGC |
| R CTAGCTAGCTCAACAGCTCACTCTGCT | |
| pCMX-GAL4-HIF1A | F CGGATATCAATGGAGGGCGCCGGCG |
| R CTAGCTAGCTCAGTTAACTTGATCCAA AGCT | |
| pCMX-VP16-P300CH1 | F CGCGGATCCTATGGCCGAGAATGTGG TGGAAC |
| R CTAGCTAGCCCAACGGGTGCTCCAGT CAAA | |
| pCDNA3.1(+)-HIF1A | F CGGGGTACCTATGGAGGGCGCCGGC |
| R TGCTCTAGATCAGTTAACTTGATCCAAAGC | |
| pCDNA3.1(+)-TFAP2C | F CGGGGTACCACGCCGGACGCCATGTTG |
| R TGCTCTAGACTCTCCTAACCTTTCTTC GTTCC | |
| PGL3basic-VEGF promoter | F GGGGTACCTTTGGGTTTTGCCAGACT |
| R CCGCTCGAGAGGAGGGAGCAGGAATAG | |
| PGL3basic-PITX2C promoter | F GGGGTACCGGGGACAAAAGGACTTTC |
| R CCGCTCGAGCCCTGTTGGCCTAACATC |
Site-directed mutagenesis and plasmid construction
Human CITED2 and HIF1A cDNA were obtained from OriGene True-Clone, and TFAP2C cDNA was purchased from GeneCopoeia. CITED2 mutations were constructed by using the Quick Change Lightning Site-Directed Mutagenesis kit (Strata gene, La Jolla, CA, USA). Then the introduced mutations were confirmed by DNA sequence.
The WT and mutant CITED2 were amplified by PCR from cDNA and inserted into the pEGFP-N1 vector (BD Biosciences, Palo Alto, CA, USA). The ORF of HIF1A and TFAP2C were also amplified by PCR from cDNA and inserted respectively into the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA, USA) to create the expression plasmid pcDNA3.1-HIF1A and pcDNA3.1-TFAP2C.
A 1300-bp fragment of the p300-CH1, PITX2C promoter and an 870-bp segment of VEGF promoter amplified by PCR from Human genomic DNA were cloned respectively into the GAL4-pCMX vector and the luciferase reporter PLG3-basic vector. GAL4-HIF1A was constructed by cloning DNA fragments into GAL4-pCMX vector at the Ecorv and Nhel sites. All primers of the PCRS were list in Table 2.
The VP16-pCMX vector with the potent transactivating domain of HSV, the promoter pGL3-basic vector with 4×GAL4 DNA-binding sites and the GAL4-pCMX vector containing GAL4-DBD were provided by Dr. Ronald M. Evans (Salk Institute for Biological Studies, USA).
Cell culture and transient transfection
293T and Hela cells were maintained in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, 100 mg/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Transfection was carried out using a standard calcium phosphate method or Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA, USA).
Subcellular localization
Hela cells were seeded in 12-well tissue culture plates 20 h prior to transfection at approximately 60% confluency. GFP-CITED2 expression constructs containing wild-type and mutant CITED2 were transfected using Lipofectamine 2000, according to the manufacturer's instructions. The empty vector pEGFP-N1 was transfected as a control. Forty hours after transfection, the cells were fixed and permeabilised in 4% paraformaldehyde for 15 min, 0.1% Triton X-100 for 20 min and the DNA was stained with 0.5 µg/ml DAPI for 3 min at room temperature. The cells were observed by fluorescence microscopy. All steps were operated in lucifugal conditions.
Mammalian two-hybrid assay and transcriptional assays
Mammalian two-hybrid assay plasmids including pCMX-VP16-p300, TK promoter reporter plasmid, the Renilla luciferase control plasmid pREP7-RLu and pCMX- GAL4-CITED2 (wild-type or mutant) or pCMX-GAL4-HIF1A were contransfected into 293T cells. Thirty hours after transfection, cells were washed and lysed in passive lysis buffer (Promega, Madison, WI, USA) and the transfection efficiency was normalised to paired Renilla luciferase activity by using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
In addition, the Dual LuciferaseReporter Assay System was used to study the effect of CITED2 on the transcription of VEGF and PITX2C. Plasmids consisting of the Renilla luciferase control plasmid pREP7-RLu, pcDNA3.1-CITED2 (wild-type or mutant), PGL3-VEGF-pro and pcDNA3.1-HIF1A or PGL3-PITX2C-pro and pcDNA3.1-TFAP2C were contransfected into 293T cells. Thirty hours after transfection, cells were treated the same way as above.
Statistical analysis
The results represent the means of three independent experiments performed in triplicate, and the bars denote the S.D. The independent-samples t test was adopted to determine statistical significance of unpaired samples. All data were analyzed by Prism Demo 5 software.
Results
Genetic and bioinformatics analysis
From a total of 700 non-syndromic CHD patients, we identified five novel CITED2 nucleotide alterations (two amino acid deletions and three amino acid substitutions, table3). Three mutations (c.C548T, c.A586G and c.574-59delAGCGGC) were found in one, one and four patients with Ventricular septal defect (VSD) respectively. One mutation (c.C418T) was detected in one patient with Tetralogy of Fallot (TOF) and another mutation (c.481–483delAGC) was detected in one patient with Artrial septal defect (ASD).
Table 3. Position of variations.
| Coding position | Amino acid position | Phenotype of mutation carrier |
| c.C418T | p. P140S,Pro-Ser | F4 |
| c.C548T | p. S183L,Ser-Leu | VSD |
| c.A586G | p. S196G,Ser-Gly | VSD |
| c.481–483delAGC | p.Ser161delAGC | ASD |
| c.574–579delAGCGGC | p.Ser192_Gly193delAGCGGC | VSD |
All potential pathogenic mutations have not been reported in the NCBI dbSNP and are not included in the 1000 Genome Project database (http://browser.1000genomes.org/).
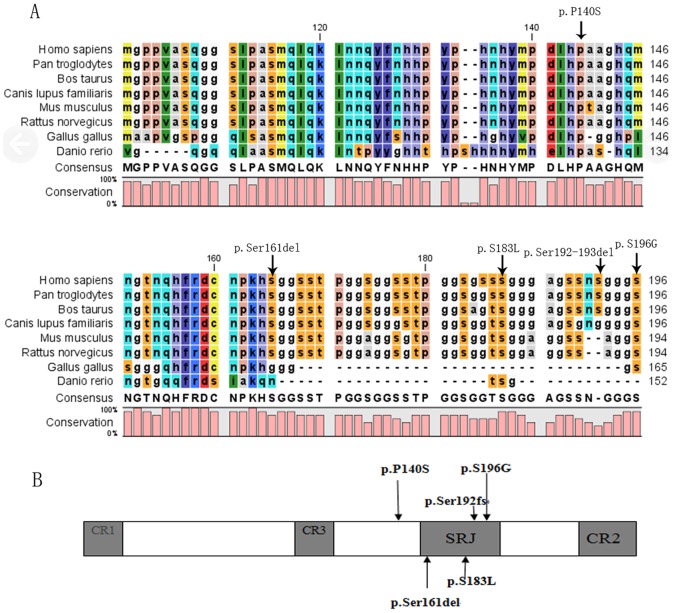
The result of sequence alignment of CITED2 proteins among several species showed that three acid substitutions were located at highly conserved regions among different species (human, chimpanzee, mice, dog, cattle, rat, chicken and zebrafish) and two amino acid deletions were not located at highly conserved regions among these species (Figure 1).
Figure 1. Structure of CITED2.
A: Sequence alignment of CITED2 proteins among several species. The figure showed that three acid substitutions were located at highly conserved regions among many species (human, chimpanzee, mice, dog, cattle, rat, chicken and zebrafish). B: Position of mutations in the CITED2 protein identified in CHD patients. CITED2 has three conserved regions CR1-3 and serine-glycine rich junction (SRJ). All other mutations were located in SRJ except p.P140S.
CITED2 mutations decrease HIF1A repression leading to up-regulation of VEGF expression
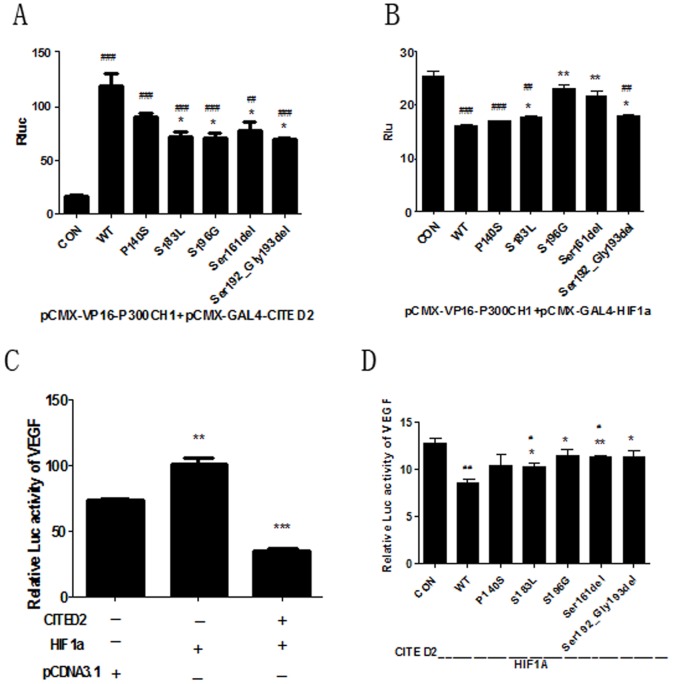
Two mammalian two-hybrid assays were used to evaluate whether the mutation affected the interaction between every two of CITED2, p300CH1 and HIF1A (Figure 2). Cotransfection of both VP16- P300 and wild-type GAL4-CITED2 with the TK promoter reporter plasmid led to a nearly 10-fold increase in luciferase activity compared with VP16-P300 and empty vector of CMX-GAL4 (t test, p<0.01). The luciferase activity of p. P140S mutant was even the same as the wt-type, However, cotransfection of VP16-P300 and the four mutants (p.S183L, p.S196G, p.Ser161delAGC, p.Ser192_Gly193delAGCGGC) GAL4-CITED2 showed weakened luciferase activity (t test, p<0.05) (Figure 2A) compared with wt-type. These findings indicated that the four mutations diminished protein-protein interactions between p300 and CITED2, but the p.P140S mutant didn't alter the interactions.
Figure 2. Effect of CITED2 mutations on the transcriptional activation of HIF1A to its target gene VEGF.
A: Effect of mutations on CITED2-p300CH1 interactions. We cotransfected 293T cells with pCMX-VP16-p300CH1, TK promoter reporter plasmid, and the Renilla luciferase internal control plasmid, as well as empty vector pCMX-GAL4, GAL4-CITED2 wild-type, and the mutants. The significance of differences was calculated using the independent-samples t test. (*p<0.05, **p<0.01 versus. wt-type, #p<0.05, ##p<0.01 versus.empty vector pCMX-GAL4.) B: Effect of mutations on HIF1A-p300CH1 interactions. Cotransfection of pCMX-VP16-p300CH1, pCMX-GAL4-HIF1A, TK promoter reporter plasmid, and the Renilla luciferase internal control plasmid, as well as empty vector pcDNA3.1 (+), pcDNA3.1 (+)-CITED2 wild-type, and the mutant. (* p<0.05, ** p<0.01 versus wt-type, # p<0.05, ## p<0.01 versus. empty vector pcDNA3.1 (+)) C: Effect of wt-type on the transcriptional activation of VEGF. Transfected the VEGF reporter plasmid and the expression vector for HIF1A, CITED2 or pcDNA3.1 were transfected together in 293 T cells. The luciferase activity was normalized to Renilla activity.* p<0.05, **p<0.01 versus the untreated group (n = 3). D: Effect of CITED2 mutants on transcription activation of VEGF compared with CITED2-wt. The rest report plasmids were same as above. (*p<0.05, **p<0.01 versus wt-type, #p<0.05, ##p<0.01 versus empty vector pcDNA3.1(+)). The results represent the means of 3 independent experiments performed in triplicate and the significance of differences was calculated using independent-samples t test.(CITED2 = Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2, HIF1A = Hypoxia Inducible Factor 1, VEGF = vascular endothelial growth factor)
Another mammalian two-hybrid assay was operated and analyzed to further evaluate whether the repression of HIF1A - p300 complex was influenced by CITED2 mutation. The result showed that the luciferase activity of wt-type was only 60% of the control (t test, p<0.01) (Figure 2B). Compared with wild-type, the luciferase activity of mutants increased obviously except the p.P140S mutant. In conclusion, CITED2 mutations weaken the HIF1A repression by diminishing the protein-protein interactions between p300CH1 and CITED2 on the one hand and by enhancing the interactions between p300CH1 and HIF1A on the other hand.
As HIF1A can induce vascular endothelial growth factor (VEGF) potently, we supposed that CITED2 mutations influenced the transcription of VEGF through their effect on HIF1A. This was confirmed by our dual luciferase assay (Figure 2C). Wild-type CITED2 caused an approximately 32% decrease of activity compared with the control (t test, p<0.01). P140S showed no difference with wild-type in luciferase activity. As for the other four mutants, Ser161delAGCAGC showed an observable promotion of VEGF-promoter resulting in higher luciferase activity than wild-type (t test, p<0.01) and the rest mutants showed few differences compared with wt-type (t test, p<0.05) (Figure 2D).
CITED2 mutations impair TFAP2C coactivation resulting in abnormal transactivation of PITX2C
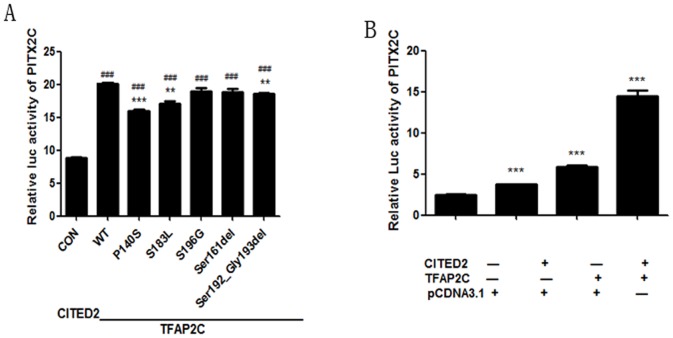
As a transcriptional coactivator of TFAP2, CITED2 influenced cardiac left-right patterning by regulating the left-right patterning Nodal-PITX2C pathway. PITX2C is a critical gene of the Nodal-PITX2C pathway and controls the location of heart and intestines in embryo. Our study showed that CITED2 mutations resulted in decreased luciferase activity of PITX2 by diminishing the coactivation of CITED2 and TFAP2C. The luciferase activity of three mutants were decreased obviously compared with wt. (p.P140S vs. wt-type 80% (t test, p<0.01), p.S183L vs. wt-type 85% (t test, p<0.01), p.Ser192_Gly193delAGCGGC vs. wt-type 92% (t test, p<0.01)) (Figure 3A). The rest two mutants coactivated TFAP2C to the same level as wt-type.
Figure 3. Effect of CITED2 variants on the cooperation between CITED2 and TFAP2C in the transactivation of the PITX2C.
A: Effect of CITED2 mutations on the transcription activation of PITX2C. (*p<0.05, **p<0.01 versus wt-type, #p<0.05, ##p<0.01 versus.empty vector pcDNA3.1(+)). B: CITED2-wt and TFAP2C working on the transcriptional activation of PITX2C. PITX2C reporter plasmid and the expression vector for TFAP2C, CITED2, or pcDNA3.1 alone were transfected respectively in 293 T cells. The luciferase activity was normalized to Renilla activity.(* p<0.05, **p<0.01 versus the untreated group (n = 3)).
In addition, we designed another test to prove the TFAP2 coactivation with CITED2. The result showed that cotransfection of empty vector of pcDNA3.1 (+) with the luciferase reporter PGL3-PITX2C-pro was the lowest in all groups including pcDNA3.1-TFAP2C or wt–type pcDNA3.1-CITED2 only and both of them (Figure 3B).
In conclusion, CITED2 mutations contributed to the abnormal transactivation of PITX2C.
Impact of CITED2 mutations on Subcellular Localization
To further study whether the functional changes are caused by changed subcellular localization of the protein, the transfections were performed using N-terminal GFP fusion constructs of wt and mutant CITED2, followed by fluorescence microscopy. The result indicated that the effects of CITED2 mutations on VEGF and PITX2C were not caused by the incorrect localization of the protein. Whether in wt or mutant of CITED2 the proteins were discovered mainly in nucleus and a lesser degree in the cytoplasm of Hela (Figure S1).
Discussion
Previous researches of cited2-/-mice confirmed that cited2 plays a critical role in the development of heart and is essential for the normal creation of the left–right axis. Cited2-/- embryos showed a series of cardiac malformations such as VSD, ASD, outflow tract abnormalities and abnormal heart looping.
We screened the coding region and splice sites of the CITED2 gene in 700 Chinese CHD patients. Two potential pathogenic amino acid deletions (p.Ser161delAGCAGC and p.Ser192_Gly193delAGCGGC) and three potential pathogenic amino acid substitutions variants (p. P140S, p. S183L and p. S196G) were identified. These three regional highly conserved substitutions (conserved among Humans, chimpanzee, mice, dog, cattle, rat, chicken and zebrafish) were not identified in control group or the variant databases. Therefore, we supposed that these three mutations were possibly causative. Since, SRJ region is a research hot spot at present, the two potential pathogenic amino acid deletions in our study were found in SRJ region. As a result, the necessity of this study is highlight. Although the CHD phenotype was not seen in SRJ-deficient mice as observed in mutation carrying patients, we supposed that this could be due to species differences [23], [24]in the function of CITED2, or some other unidentified factors[25] might interact with CITED2 and modify its phenotype. Alternatively, it is also possible that CHD were present earlier in life but spontaneously closed at a later time in SRJ-deficient mice.
Mammalian two-hybrid analysis permits the semi-quantitative assessment of protein-protein interactions occurring within living cells. Cotransfection of wt or mutant CITED2 and p300CH1 in 293Tcells, the binding between CITED2 and p300CH1 activated the TK report gene expression in vivo. The functional study greatly supported the hypothesis that the mutations are causative and might affect the formation of heart. The last four mutated proteins (p. S183L, p. S196G, p.Ser161delAGCAGC and p.Ser192_Gly193delAGCGGC) showed significantly decreased reporter gene activation ability compared with wt-type. However, an opposite phenomenon occurred by transfecting p300CH1, HIF1A and wt or mutant CITED2 together in cells. Taken together,the results indicated that the four mutated proteins decreased the interaction between CITED2 and p300CH1 compared with wt-type,causing a weakened competitive binding to p300 CH1 of CITED2. The increased interaction between HIF1A and p300CH1 could up- regulate the promoter activity of VEGF according to our dual luciferase experiment.
Our study also showed that three mutations decreased the consociation of TFAP2C and CITED2 in the transactivation of pitx2c, an essential gene of the left–right axis establishment confirmed in mice and chick embryo. The mice experiments already indicated that knocking out pitx2c gene can lead to valve defects, body wall dysraphism, gastroschisis, ectopia cordis and other multiple organs polymorphous defects. In addition, there was no evidence that CITED2 mutations were involved in the incorrect location of the protein in the subcellular localization experiment.
In conclusion, we identified five novel mutations among 700 CHD patients by screening the coding region and splice sites of the CITED2 gene. To confirm our hypothesis that the mutations were pathogenic, we investigated the function and mechanism of them. Our study revealed that four mutations influenced the transcription regulatory properties of VEGF and three mutations reduced costimulation capacity to promote PITX2C. Further research showed that four CITED2 mutations recovered the promoter activity of VEGF [26]caused by its decreased competitiveness with HIF1A to bind the p300CH1. Furthermore, three mutations also decreased the consociation of TFAP2C and CITED2 in the transactivation of PITX2C. Our study confirmed that CITED2 is a disease-causing gene of CHD and its mutations can result in the cardiac malformations.
Supporting Information
Subcellular localization of CITED2. Localization of wild-type and mutant CITED2 GFP-fusion protein in transfected Hela cells were observed by fluorescent microscope. The empty vector pEGFP-N1 was transfected as a control. All figures were drawn by fluorescence microscopy and Adobe Photoshop CS5.
(TIF)
Funding Statement
This work was supported by the National Basic Research Program of China (2010CB529504), the National Natural Science Foundation of China (81300131) and the Applied Basic Research Program of Qinghai Province (QH2013-z-744). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoffman JI, Kaplan S (2002) The incidence of congenital heart disease. J Am Coll Cardiol 39: 1890–1900. [DOI] [PubMed] [Google Scholar]
- 2. Crider KS, Bailey LB (2011) Defying birth defects through diet? Genome Med 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blue GM, Kirk EP, Sholler GF, Harvey RP, Winlaw DS (2012) Congenital heart disease: current knowledge about causes and inheritance. Med J Aust 197: 155–159. [DOI] [PubMed] [Google Scholar]
- 4. Xiong F, Li Q, Zhang C, Chen Y, Li P, et al. (2013) Analyses of GATA4, NKX2.5, and TFAP2B genes in subjects from southern China with sporadic congenital heart disease. Cardiovasc Pathol 22: 141–145. [DOI] [PubMed] [Google Scholar]
- 5. Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, et al. (2005) Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet 37: 423–428. [DOI] [PubMed] [Google Scholar]
- 6. Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, et al. (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447. [DOI] [PubMed] [Google Scholar]
- 7. Andrews JE, O'Neill MJ, Binder M, Shioda T, Sinclair AH (2000) Isolation and expression of a novel member of the CITED family. Mech Dev 95: 305–308. [DOI] [PubMed] [Google Scholar]
- 8. Li Q, Pan H, Guan L, Su D, Ma X (2012) CITED2 mutation links congenital heart defects to dysregulation of the cardiac gene VEGF and PITX2C expression. Biochem Biophys Res Commun 423: 895–899. [DOI] [PubMed] [Google Scholar]
- 9. Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, et al. (2002) The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A 99: 10488–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu M, Wu X, Li Y, Yang X, Hu J, et al. (2014) CITED2 mutation and methylation in children with congenital heart disease. J Biomed Sci 21: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, et al. (2003) Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol 10: 504–512. [DOI] [PubMed] [Google Scholar]
- 12. Amati F, Diano L, Campagnolo L, Vecchione L, Cipollone D, et al. (2010) Hif1alpha down-regulation is associated with transposition of great arteries in mice treated with a retinoic acid antagonist. BMC Genomics 11: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macdonald ST, Bamforth SD, Braganca J, Chen CM, Broadbent C, et al. (2013) A cell-autonomous role of Cited2 in controlling myocardial and coronary vascular development. Eur Heart J 34: 2557–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu B, Doughman Y, Turakhia M, Jiang W, Landsettle CE, et al. (2007) Partial rescue of defects in Cited2-deficient embryos by HIF-1alpha heterozygosity. Dev Biol 301: 130–140. [DOI] [PubMed] [Google Scholar]
- 15. Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, et al. (2001) Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29: 469–474. [DOI] [PubMed] [Google Scholar]
- 16. Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, et al. (2000) Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet 25: 42–46. [DOI] [PubMed] [Google Scholar]
- 17. Bhattacherjee V, Horn KH, Singh S, Webb CL, Pisano MM, et al. (2009) CBP/p300 and associated transcriptional co-activators exhibit distinct expression patterns during murine craniofacial and neural tube development. Int J Dev Biol 53: 1097–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, et al. (2001) Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol 231: 252–264. [DOI] [PubMed] [Google Scholar]
- 19. Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, et al. (2004) Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet 36: 1189–1196. [DOI] [PubMed] [Google Scholar]
- 20. Sperling S, Grimm CH, Dunkel I, Mebus S, Sperling HP, et al. (2005) Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum Mutat 26: 575–582. [DOI] [PubMed] [Google Scholar]
- 21. Yang XF, Wu XY, Li M, Li YG, Dai JT, et al. (2010) [Mutation analysis of Cited2 in patients with congenital heart disease]. Zhonghua Er Ke Za Zhi 48: 293–296. [PubMed] [Google Scholar]
- 22. Chen CM, Bentham J, Cosgrove C, Braganca J, Cuenda A, et al. (2012) Functional significance of SRJ domain mutations in CITED2. PLoS One 7: e46256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li DY, Whitehead KJ (2010) Evaluating strategies for the treatment of cerebral cavernous malformations. Stroke 41: S92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruiz-Perez VL, Blair HJ, Rodriguez-Andres ME, Blanco MJ, Wilson A, et al. (2007) Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development 134: 2903–2912. [DOI] [PubMed] [Google Scholar]
- 25. Bentham J, Michell AC, Lockstone H, Andrew D, Schneider JE, et al. (2010) Maternal high-fat diet interacts with embryonic Cited2 genotype to reduce Pitx2c expression and enhance penetrance of left-right patterning defects. Hum Mol Genet 19: 3394–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agrawal A, Gajghate S, Smith H, Anderson DG, Albert TJ, et al. (2008) Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum 58: 3798–3808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Subcellular localization of CITED2. Localization of wild-type and mutant CITED2 GFP-fusion protein in transfected Hela cells were observed by fluorescent microscope. The empty vector pEGFP-N1 was transfected as a control. All figures were drawn by fluorescence microscopy and Adobe Photoshop CS5.
(TIF)