Figure 1. Comparison of adaptive and static T-ReX simulations for the refinement of protein target decoys 1kviA, 1pgx, 1cy5A, 1shfA, 1r69, 1csp, 1ah9 and 1b72A.
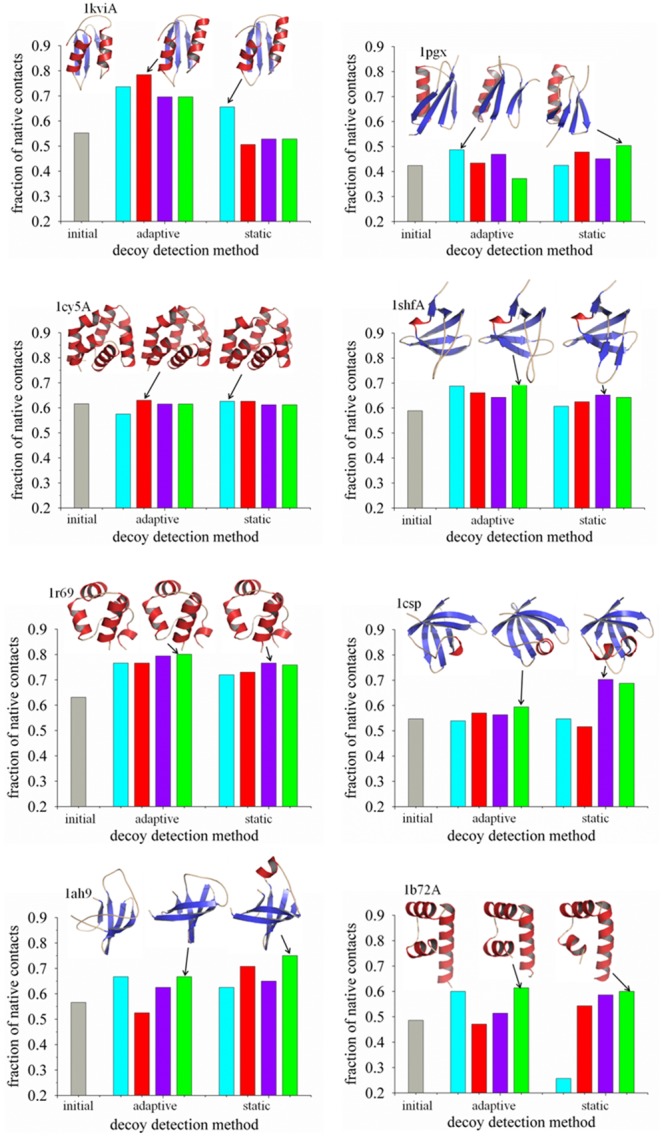
A bar graph is shown of the computed fraction of native contacts for each detection method. Conformers were culled from the ensemble of replica clients at a temperature of 275-order rank detection is based on the energy functions CHARMM22/GBMV2 (cyan-colored bar), GOAP (red bar), dDFIRE (purple bar), and RWplus (green bar). For each target, the initial unrefined decoy is the conformer with the highest fraction of native contacts (grey bar). With the exception of GOAP scoring, refinement results using the static method for 1kivA, 1pgx, 1cy5A, 1shfA, 1r69 and 1b72A were taken from an earlier reported study.5 Molecular models are illustrated for the crystallographic structure and the top-ranked conformers from the adaptive and static sampling methods.
