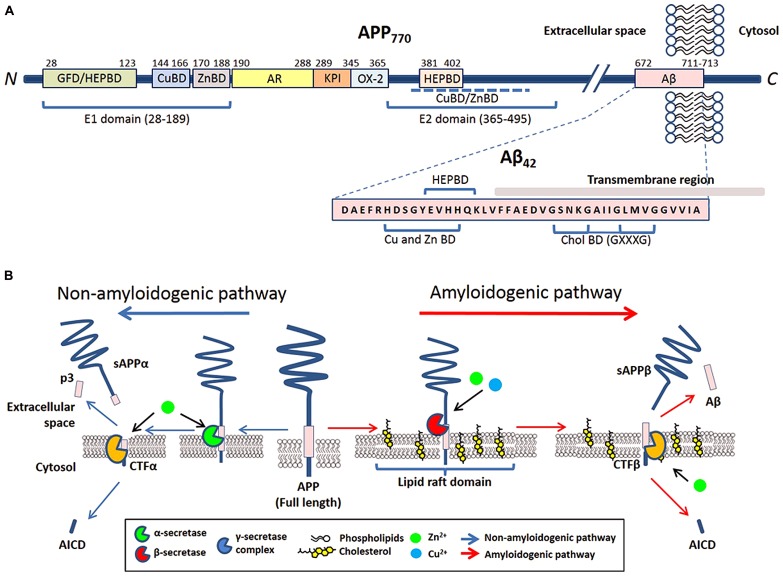
FIGURE 1.
The involvement of metals and cholesterol in post-translational modification of APP. (A) Schematic of reported metal and cholesterol binding domains in APP770 and Aβ in relation to other recognized motifs. APP770 is the longest isoform of APP with the APP751 isoform lacking the OX-2 domain and the neuron prevalent isoform APP695 lacking both OX-2 and Kunitz-type protease inhibitor (KPI). Within the extracellular presented ectodomain of APP, the E1 region at the N-terminal contains a copper binding domain (CuBD) and zinc binding domain (ZnBD) that is C-terminally orientated compared to the growth factor domain (GFD) which incorporates a heparin binding domain (HEPBD). The E1 domain is followed by the acidic region (AR), KPI and OX-2 before the E2 domain of APP, containing a HEPBD and CuBD/ZnBD that is yet to be exactly mapped (Dahms et al., 2012). The E2 domain is followed by the Aβ peptide that is partially embedded into the transmembrane region. Aβ also has a recognized CuBD/ZnBD as well as a Cholesterol binding region (CholBD) that incorporates the GXXXG motifs. (B) Proteolytic processing of APP predominantly follows two pathways that are initiated by separate secretases. The non-amyloidogenic pathway (blue arrows) initiates with the cleavage of full-length APP by α-secretase within the Aβ sequence. Following further cleavage by the γ-secretase complex, this pathway results in the generation of soluble N-terminal APP fragment (sAPPα) and C-terminal fragments (p3 and AICD). The alternative amyloidogenic pathway (red arrows) involves sequential cleavage of APP by β-secretase followed by the γ-secretase complex, which results in the liberation of a soluble N-terminal sAPPβ fragment, Aβ peptide, and AICD. Copper and zinc affect the processing of APP and Aβ generation on neuronal membranes through their direct influence on the enzymatic activity of β-, α-, and γ-secretases. The influence of cholesterol is through its requirement in lipid raft domains, the location for amyloidogenic processing of APP.