Table 1.
Electron transport chain complexes and regulated genes
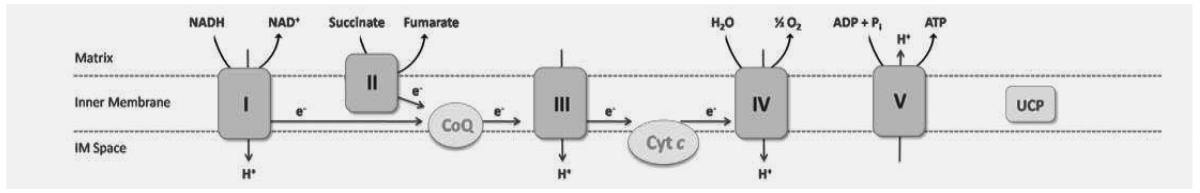
| |||||||
|---|---|---|---|---|---|---|---|
| Complex I NADH Dehydrogenase |
Complex II Succinate Dehydrogenase |
Complex III Cytochrome c Reductase |
Complex IV Cytochrome c Oxidase |
Complex V F0F1 ATPase |
UCP Uncoupling Protein |
Chaperone | |
|
| |||||||
| Mitochondrial genome encoded subunits |
7 | 0 | 1 | 3 | 2 | 0 | 0 |
| Nuclear genome encoded subunits |
40 | 4 | 10 | 15 | 19 | 5 | 3 |
|
| |||||||
| Number of differentially regulated genesa |
26 | - | 8 | 8 | 9 | 2 | 1 |
|
| |||||||
| Most temporally regulatedb |
Ndufb6 | - | Uqcrc1 | Cox4i1 | Atpaf1 | Ucp2 | Sco1 |
| (Fold changed)c | ▼ 9.1 | ▲ 3.2 | ▲ 4.0 | ▲ 5.3 | ▲ 3.1 | ▼ 1.64 | |
|
| |||||||
| Dose-responsived | Ndufa10 | - | Cyc1 | Cox7b | Atp5g3 | Ucp2 | Sco1 |
| Atp5l | Ucp5 | ||||||
|
| |||||||
Where |fold change| > 1.5 and P-value < 0.1 in the time course experiment
Gene exhibiting maximum temporal fold change across the time course experiment with P-value < 0.1 in the given ETC complex
Maximum temporal fold change across the time course experiment with P-value < 0.1
Gene exhibiting dose-response to TCDD in the given ETC complex
