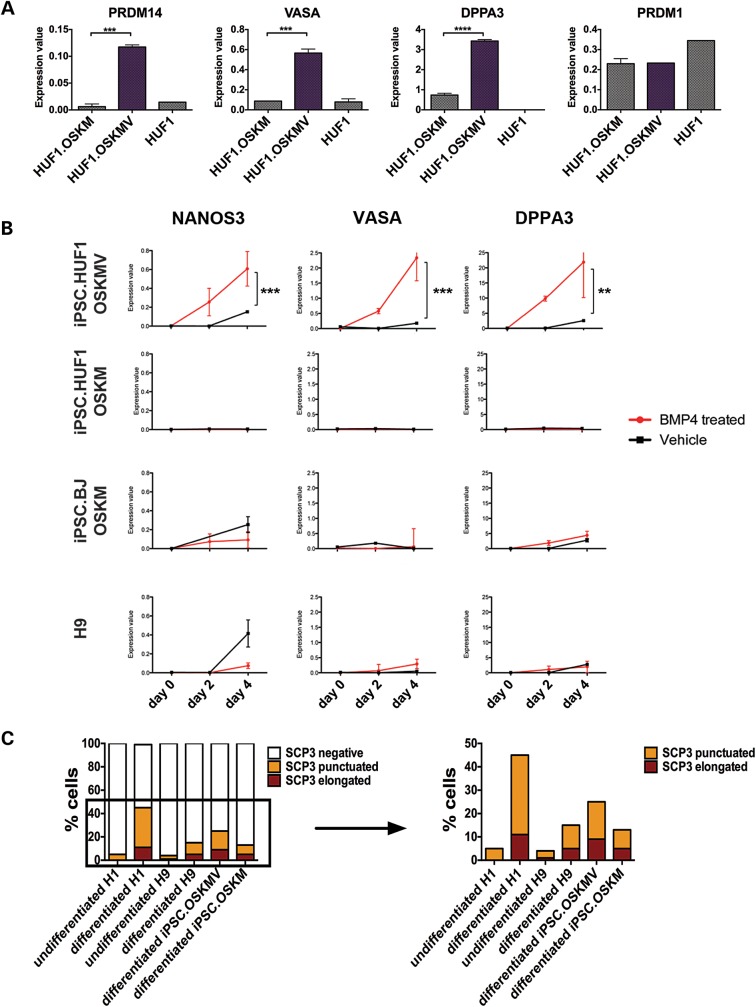
Figure 2.
Functional and molecular studies of iPSC.HUF1 derived with OSKM and OSKMV. (A) Gene expression analysis of markers associated with the germline lineage in an undifferentiated state. Three genes (PRDM14, VASA, DPPA3) showed significantly higher expression in OSKMV-derived clones (purple bar) compared with OSKM (Student's t-test, mean ± SD; n ≥ 16 for each gene and sample *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (B) Gene expression analysis of clones derived with OSKM and OSKMV, iPSC.BJ line and H9 during in vitro differentiation into primordial germ cells with BMP4. Samples were isolated at day 0, 2 and 4 post-BMP4/vehicle treatment. Three key markers (NANOS3, VASA, DPPA3) were upregulated in OSKMV-derived clones (Student's t-test, mean ± SD; n ≥ 16 for each gene and sample *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (C) Quantification of meiotic spreads. Approximately 200–300 cells were counted for each line. Staining patterns were classified as negative, punctuated or elongated.