Abstract
Background and Aims
Forisomes are specialized structural phloem proteins that mediate sieve element occlusion after wounding exclusively in papilionoid legumes, but most studies of forisome structure and function have focused on the Old World clade rather than the early lineages. A comprehensive phylogenetic, molecular, structural and functional analysis of forisomes from species covering a broad spectrum of the papilionoid legumes was therefore carried out, including the first analysis of Dipteryx panamensis forisomes, representing the earliest branch of the Papilionoideae lineage. The aim was to study the molecular, structural and functional conservation among forisomes from different tribes and to establish the roles of individual forisome subunits.
Methods
Sequence analysis and bioinformatics were combined with structural and functional analysis of native forisomes and artificial forisome-like protein bodies, the latter produced by expressing forisome genes from different legumes in a heterologous background. The structure of these bodies was analysed using a combination of confocal laser scanning microscopy (CLSM), scanning electron microscopy (SEM) and transmission electron microscopy (TEM), and the function of individual subunits was examined by combinatorial expression, micromanipulation and light microscopy.
Key Results
Dipteryx panamensis native forisomes and homomeric protein bodies assembled from the single sieve element occlusion by forisome (SEO-F) subunit identified in this species were structurally and functionally similar to forisomes from the Old World clade. In contrast, homomeric protein bodies assembled from individual SEO-F subunits from Old World species yielded artificial forisomes differing in proportion to their native counterparts, suggesting that multiple SEO-F proteins are required for forisome assembly in these plants. Structural differences between Medicago truncatula native forisomes, homomeric protein bodies and heteromeric bodies containing all possible subunit combinations suggested that combinations of SEO-F proteins may fine-tune the geometric proportions and reactivity of forisomes.
Conclusions
It is concluded that forisome structure and function have been strongly conserved during evolution and that species-dependent subsets of SEO-F proteins may have evolved to fine-tune the structure of native forisomes.
Keywords: Forisome evolution, Dipteryx panamensis, Medicago truncatula, Pisum sativum, Vicia faba, Lotus japonicus, Canavalia gladiata, macromolecular assembly, papilionoid legumes, Papilionoideae, Fabaceae, sieve element occlusion gene family, SEO, structural phloem protein
INTRODUCTION
The phloem tissue of vascular plants contains a sieve tube system for the transport of photoassimilates, comprising longitudinally aligned cells known as sieve elements, which are connected by perforated sieve plates (Esau, 1969). During maturation, sieve elements undergo selective autolysis to produce metabolically inactive mature sieve elements that retain only a few organelles as well as specialized structural phloem proteins (P-proteins) (Esau and Cronshaw, 1967). The P-proteins are normally located around the periphery of the sieve element, thus facilitating mass flow through the phloem (Knoblauch and van Bel, 1998).
When the phloem is wounded, P-proteins detach from their parietal location and prevent the loss of photoassimilates by plugging the sieve plates (Anderson and Cronshaw, 1970; Knoblauch and van Bel, 1998; Ernst et al., 2012). Strasburger (1891) discovered a special type of crystalloid P-protein in Robinia pseudoacacia, and such proteins appear to be exclusive to the papilionoid legumes (Behnke, 1981; Peters et al., 2010). They were named forisomes (from the Latin foris meaning gate and the Greek soma meaning body) because they undergo a reversible, anisotropic conformational change from a condensed spindle-like shape to an expanded/dispersed plug-like state, allowing them to block the sieve plates and act as a ‘gatekeeper’ (Knoblauch et al., 2003). The reaction is triggered by mechanical or physical phloem injuries that cause either the direct influx of calcium ions, or electropotential waves that propagate the signal over long distances and promote the local influx of calcium ions into the sieve elements (Knoblauch et al., 2003; Furch et al., 2007; Hafke et al., 2009). This in turn triggers a rapid, ATP-independent conformational change and thereby enables forisomes to regulate the pressure-driven mass flow of assimilates by occluding the sieve tubes (Knoblauch et al., 2001, 2003). During this conformational change, the longitudinal contraction and radial expansion of the forisome causes an increase in volume that is probably mediated by an influx of water into the forisome body (Pickard et al., 2006; Schwan et al., 2009). A 3- to 9-fold volume increase occurs in vitro, although this is influenced by the preparation method (Knoblauch et al., 2003; Schwan et al., 2007; Peters et al., 2007, 2008; Knoblauch et al., 2012). In addition to Ca2+ and other divalent cations, non-physiological pH values can also induce conformational change in forisomes (Knoblauch et al., 2003; Schwan et al., 2009; Müller et al., 2010). Isolated forisomes remain fully functional in the absence of dissolved oxygen (Schwan et al., 2007) and more than 5000 contraction–expansion cycles have been induced by electrotitration (Jäger et al., 2008) thereby generating forces of up to 0·1 μN per forisome in each cycle (Schwan et al., 2009).
The structure of forisomes has been investigated by transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (Wergin and Newcomb, 1970; Palevitz and Newcomb, 1971; Esau, 1978; Lawton, 1978a, b; Schwan et al., 2007; Jäger et al., 2008). These experiments revealed that the smallest detectable units are filaments, 3–5 nm in diameter, which assemble into fibrils (Arsanto, 1982; Groscurth et al., 2012). The protein domains responsible for such protein–protein interactions are not yet understood, although helical structures may be involved (Lawton, 1978a; Arsanto, 1982; Groscurth et al., 2012). The fibrils then assemble into fibres, 400–700 nm in diameter (Jäger et al., 2008), which gather into the typical spindle shape (Groscurth et al., 2012). Forisomes in some species feature tail-like protrusions (Mrazek, 1910; Lawton, 1978a), but their occurrence does not appear to follow a strict evolutionary pattern (Peters et al., 2010).
The Fabaceae family of plants evolved approximately 59 million years ago and divided rapidly into the subfamilies Caesalpinioideae, Mimosoideae and Papilionoideae (Lavin et al., 2005). Forisomes are only present in the Papilionoideae (Peters et al., 2010), which includes more than 13 000 species (Lewis et al., 2005). The subfamily Papilionoideae is divided further into the ‘basal papilionoids’, genistoids, dalbergoids, indigoferoids, milletioids, robinoids, the inverted repeat-lacking clade (IRLC), characterized by the absence of a large inverted repeat in the chloroplast genome, and a number of smaller clades (Pennington et al., 2001; Wojciechowski et al., 2004; Lavin et al., 2005; Lewis et al., 2005; McMahon and Sanderson, 2006; Bello et al., 2009; LPWG, 2013). Forisome ultrastructure has been studied extensively by TEM, but investigations in the 1970s and 1980s focused mainly on species such as Phaseolus vulgaris, which appeared approximately 8 million years ago (Palevitz and Newcomb, 1971; Esau, 1978; Lawton, 1978a, b; Lavin et al., 2005). The examined species are members of the Old World clade, comprising the mirbeloids, baphioids, indigoferoids, milletioids, robinoids and IRLC (Lavin et al., 2005; LPWG, 2013).
There are no molecular and only a few structural data on forisomes (e.g. Swartzia simplex and Bobgunnia madagascariensis) from Papilionoideae species available that belong to lineages with basal branching points in the phylogenetic tree of Papilionoideae (Behnke, 1981; Lavin et al., 2005; Peters et al., 2010; LPGW, 2013). We therefore carried out the first comprehensive analysis of forisomes from Dipteryx panamensis, a tropical tree native to Costa Rica representing the basal monophyletic tribe Dipterygeae (Pennington et al., 2001). We compared forisomes from D. panamensis with those from species of the Old World clade, i.e. Canavalia gladiata, Lotus japonicus and Medicago truncatula, which diverged from a common ancestor more than 50 million years ago, as well as Pisum sativum and Vicia faba, which diverged from a common ancestor approximately 18 million years ago (Lavin et al., 2005).
Recently, a comprehensive analysis of the sieve element occlusion (SEO) gene family showed that forisomes are encoded by SEO-F (sieve element occlusion by forisome) genes, which evolved from SEO genes encoding conventional P-proteins (Rüping et al., 2010; Ernst et al., 2012). We identified additional SEO-F genes in the current investigation, including a single SEO-F gene in the basal species D. panamensis. We confirmed that forisome-like structures assembled following the heterologous expression of this gene in Nicotiana benthamiana epidermal cells. We used this background to compare native and artificial (homomeric) forisomes from several species representing a broad range of papilionoid clades, and studied forisome assembly, ultrastructure and activity by TEM, SEM and confocal laser scanning microscopy (CLSM).
MATERIALS AND METHODS
Plant material and cultivation
Canavalia gladiata, Lotus japonicus, Medicago truncatula, Pisum sativum and Vicia faba plants were cultivated in the greenhouse at 25°C with a 16-h photoperiod (light intensity 150 μmol m−2 s−1). Nicotiana benthamiana plants were cultivated in a growth chamber at 22°C with a 16-h photoperiod (light intensity 200 μmol m−2 s−1). Dipteryx panamensis was grown in a greenhouse in San José, Costa Rica.
Identification of DpSEO-F1 and LjSEO-F1
Degenerate oligonucleotide primers for the amplification of SEO genes were designed based on sequence alignments of SEO-F genes as described by Rüping et al. (2010) and were used to identify putative SEO gene coding sequences from D. panamensis. MtSEO-F1 (accession number EU016204.1) was used as a BLASTN query (Altschul et al., 1990) to identify further potential SEO sequences. MtSEO-F1 aligned with three L. japonicus cDNA clones (Sato et al., 2008), and the most strongly conserved sequence (maximum identity 81 %) was used to design appropriate oligonucleotide primers. L. japonicus and D. panamensis RNA extraction and cDNA synthesis were carried out as described by Rüping et al. (2010) and the SEO coding sequences were amplified. The 5′ and 3′ ends of both SEO sequences were identified by the rapid amplification of cDNA ends method (Frohman et al., 1988). SEO coding sequences have been deposited in GenBank (DpSEO-F1, accession number KJ439757; LjSEO-F1, accession number KJ439756).
Identification of genomic DpSEO-F1 sequence
DNA was extracted from D. panamensis as described by Cota-Sánchez et al. (2006) and the oligonucleotide primers previously used for cDNA cloning were used with additional internal primers to identify the DpSEO-F1 genomic sequence.
Co-expression constructs
The co-expression of MtSEO-F genes was achieved using combinations of two expression cassettes introduced into pBIN19 (Bevan, 1984). Genes were expressed under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and terminator. The cloning strategy for the co-expression of two or three MtSEO genes, or the co-expression of MtSEO-F2 along with SEO-F genes from other species, is described in Supplementary Data Methods.
Agroinfiltration
Vectors containing the M. truncatula forisome genes were constructed previously (Müller et al., 2010). The remaining SEO-F genes were cloned in vector pENTR4™ as described in Supplementary Data Methods. The pENTR4™–SEO constructs were introduced into the pBatTL and pBatTL–venus–ccdB vectors and transiently expressed in N. benthamiana cells as previously described (Müller et al., 2010; Groscurth et al., 2012).
Root transformation
Root transformation was carried out using binary vectors containing MtSEO-F genes fused to a humanized Renilla reniformis green fluorescent protein (hrGFP) tag under the control of the corresponding MtSEO-F promoter. The cloning strategy is described in Supplementary Data Methods. The constructs were introduced into M. truncatula using the ex vitro composite plant induction method (Collier et al., 2005) with minor modifications as described by Pélissier et al. (2008).
Confocal laser scanning microscopy
Native forisomes were analysed in stem tissue incubated in EDTA buffer (10 mm Tris–HCl pH 7·3, 10 mm EDTA, 100 mm KCl) to maintain forisomes in their condensed confirmation. For native and artificial forisomes, bright-field images were obtained by activating the transmission photomultiplier tube of the microscope. Visualization of the tails of D. panamensis and L. japonicus native forisomes was improved by incubating the corresponding stem sections for 5 min with 1 mg ml–1 sulphorhodamine in EDTA buffer and 50 % DMSO. After three washes in EDTA buffer, sulphorhodamine fluorescence was detected at an excitation wavelength of 586 nm and an emission range of 600–620 nm. Transgenic M. truncatula roots were analysed by CLSM 5 weeks after incubation with Agrobacterium rhizogenes. Roots were cut and bathed for 10 min in EDTA buffer, and hrGFP fluorescence was detected at an excitation wavelength of 488 nm and an emission range of 500–580 nm.
Ex vivo functional analysis of artificial and native forisomes
Artificial forisomes were isolated from N. benthamiana protoplasts as described by Müller et al. (2010). Native forisomes were isolated from 6-week-old M. truncatula plants. The stem rinds were peeled off and incubated for 1 h in EDTA buffer. The inner side was then scraped with a scalpel and incubated in EDTA buffer for 30 min. Plant material was ground under liquid nitrogen in a bead mill (two 1-min pulses, 30 Hz) and the ground material was resuspended in EDTA buffer before passing through a 20-μm mesh filter. The filtrate was centrifuged for 10 min at 100 g, washed in EDTA buffer and resuspended in 100 μl EDTA buffer. Forisome conformational changes were then monitored as described by Müller et al. (2010). The length and width of each forisome were measured using Gimp2 software, and the volume increase was calculated as described by Noll et al. (2011). Single fibres from purified MtSEO-F1 + MtSEO-F4 protein bodies were obtained by incubating the forisomes on a vortex mixer for 5 min before measuring their size.
Measuring the geometric parameters of forisomes
Native and artificial forisomes were monitored by CLSM and the length and width of 15 individual forisomes were measured using the Leica imaging software LAS AF. For the analysis of native forisomes, stem phloem from 6-week-old plants was incubated in EDTA buffer for 10 min. The widths of corresponding sieve plates were also determined. Artificial forisomes from N. benthamiana leaves were analysed 4 days post-infiltration. The length (l) and width (w) were determined for n = 15 native or artificial forisomes. The ratio (r) for each forisome was calculated by dividing the length by the width (l/w). Mean values and standard deviations for length, width and ratio were then calculated for each construct and species. The mean ratio therefore does not necessarily equal the mean length divided by the mean width. The geometrical data were visualized more clearly using linear graphs running through the coordinates of the mean values of length and width, and using the mean value of the ratio as the slope. The range of the graph was also adjusted to the standard deviations of length and width. For each species, we also calculated the correlation coefficient for the measured sieve plate width (spw) and forisome width (w) to test the dependence of both values. The ratio v = spw/w was calculated and compared by ANOVA (Newman–Keuls method) using SigmaPlot (Systat Software, San Jose, CA) to determine a significant difference in the group means of each species. The value of spw was determined for n = 15 in all species except C. gladiata (n = 10).
Scanning electron microscopy
Artificial forisomes were isolated as described by Müller et al. (2010) and sample preparation was carried out as described by Schwan et al. (2009). SEM images were obtained using a cold field-emission SEM (JSM-6700F, JEOL, Tokyo, Japan) and field-emission SEM (Quanta™ 3D FEG, FEI, USA) at 5 kV.
Transmission electron microscopy
Freshly prepared stem sections were incubated in EDTA buffer for 10 min prior to the fixation of native forisomes. Artificial condensed forisomes were isolated directly from sections of infiltrated N. benthamiana leaves, or the sections were first incubated in buffer containing Ca2+ (10 mm Tris–HCl pH 7·3, 100 μm CaCl2, 100 mm KCl) to promote forisome expansion. In each case, the plant material was fixed with 2 % glutaraldehyde and 2·5 % paraformaldehyde in 0·1 m phosphate buffer (pH 7·3) and 3·5 % sucrose for 3–24 h. After three washes with phosphate buffer, the samples were post-fixed by incubation with 1 % OsO4 in 0·1 m phosphate buffer (pH 7·3) for 1–2 h. The fixed samples were washed three times with distilled water and dehydrated through an ethanol series. Tissue sections were embedded in LR White Resin (Sigma-Aldrich, Germany). Thin sections were collected on copper grids and incubated with 2 % uranyl acetate for 10−30 min and lead citrate (Hanaichi et al., 1986) for 6−12 min. The sections were analysed using a Hitachi H-7100 (Hitachi, Tokyo, Japan) or a Zeiss EM900 (Carl Zeiss Microscopy, Germany) at 80 or 100 kV. The widths of protein filaments and fibrils in TEM micrographs were determined using Gimp2 software. More than ten TEM images were captured for each combination. Three independent TEM images were used for the measurement of fibril width and fine/coarse cross-striation.
Phylogenetic analysis
The phylogenetic species tree was based on that described by LPWG (2013) and visualized using Inkscape (Gould et al., 2003). The SEO protein sequences described by Rüping et al. (2010) were aligned with DpSEO-F1 and LjSEO-F1 using MUSCLE (Edgar, 2004). Gblocks (Castresana, 2000) was used to eliminate poorly aligned regions and a phylogenetic tree was created using FastTree2 (Price et al., 2010). The phylogram was visualized using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/). Pairwise amino acid sequence alignments were obtained using EMBOSS Needle (http://www.ebi.ac.uk/Tools/psa/emboss_needle/). The N-terminal (PB013523) and C-terminal domains (PB006891) were identified using the Pfam 27·0 database from the Wellcome Trust Sanger Institute (Punta et al., 2012). Thioredoxin domains were identified as previously described by Rüping et al. (2010). Domain organization and exon–intron structure were visualized using Inkscape.
RESULTS
Structural analysis of forisomes in papilionoid legumes
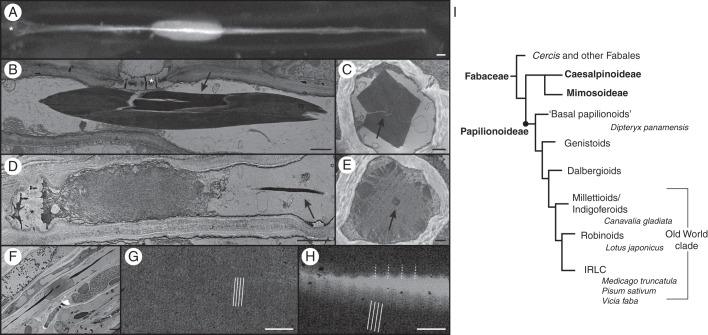
The tropical tree D. panamensis was chosen for the comprehensive structural and molecular analysis of forisomes from a basal Papilionoideae species because the presence of forisomes in this tribe has not been previously reported. Bright-field microscopy identified forisomes in the sieve elements and revealed the typical spindle shape, as described for other species, as well as tail-like protrusions at both ends. Staining with sulphorhodamine 101 indicated that the tails pervaded the entire forisome and were approximately four times the length of the main body (Fig. 1A). TEM revealed that condensed forisomes comprised an electron-dense, tightly packed protein body (Fig. 1B) composed of filaments 2·5–4·5 nm in diameter. Cross-sections of condensed D. panamensis forisomes showed that the centre of the main body was separated from the tail by a thin cleft with low electron density (Fig. 1C). The cleft may represent the physical separation of the forisome tail and main body, but we cannot exclude the possibility that the cleft is an artefact caused by specimen shrinkage during TEM preparation.
Fig. 1.
Evolution of forisomes in the Fabaceae family and structural analysis of forisomes from D. panamensis. (A) In vivo CLSM image of a D. panamensis forisome stained with sulphorhodamine 101. The tail, clearly visible as a bright streak in the image, pervades the main body of the forisome, which is revealed by lower-intensity fluorescence. (B–H) TEM of D. panamensis forisomes. (B) A condensed forisome in a mature sieve element. (C) Cross-section of a condensed forisome, indicating its typical cuboid shape. The tail is shown in the centre (arrow). (D) Expanded forisome located close to the sieve plate in a mature sieve element. The tail remains condensed (arrow). (E) Cross-section of an expanded forisome. The outline of the protein body is not yet expanded. The condensed tail is visible in the centre (arrow). (F) Expanded forisome with the condensed tail pervading the main body. (G) Magnification of the main body of a condensed forisome, showing fine 10-nm cross-striations (indicated by white lines). (H) Magnification of the tail reveals coarse cross-striations with a periodicity of 40 nm (indicated by white dotted lines), overlaid by a fine 10-nm cross-striation (indicated by white lines). (I) Simplified Fabaceae phylogenetic tree (modified from LPWG, 2013) showing the subfamilies Caesalpinioideae, Mimosoideae and Papilionoideae, the major Papilionoideae clades and the species we investigated. Smaller clades are not shown. Forisomes are only present in the Papilionoideae. The ‘basal papilionoids’ represent the first branching papilionoid lineages (Pennington et al., 2001). Branch lengths do not reflect phylogenetic distances. Sieve plates are marked with an asterisk. Scale bars: (A, B, D, F) = 2 μm, (C, E) = 500 nm, (G, H) = 100 nm.
In tissues that were wounded by cutting prior to TEM analysis, the volume of the main body increased due to the disintegration of fibrils, and forisomes were predominantly found as plugs on the sieve plates, confirming that D. panamensis forisomes are reactive (Fig. 1D). The average filament diameter in the expanded forisomes was 4–6 nm. Following forisome expansion, the sieve element lumen was completely filled with proteinaceous material, showing the ability of D. panamensis forisomes to control mass flow. In contrast, the tails always remained condensed during forisome expansion (Fig. 1E, F), as previously described for Swartzia species (Behnke, 1981; Peters et al., 2010) and evolutionarily more recent species (Wergin and Newcomb, 1970; Palevitz and Newcomb, 1971; Lawton, 1978b).
Higher magnifications of longitudinal sections of the condensed forisomes revealed fine cross-striation with a 10-nm periodicity, perpendicular to the protein body (Fig. 1G). In the tails, coarse cross-striation of 40 nm was clearly visible (Fig. 1H), but this was overlaid with finer cross-striation with similar periodicity to the cross-striations in the main body. The cross-striation patterns observed in D. panamensis forisomes were similar to those described in evolutionarily more recent species; Coronilla varia forisomes, for example, show cross-striations in the main body with 12-nm periodicity, compared with 35–40 nm in the tails (Palevitz and Newcomb, 1971; Lawton 1978b).
The proportions of forisomes from D. panamensis were then compared with those of five other species representing diverse papilionoid clades. C. gladiata was chosen to represent the milletioids, L. japonicus represented the robinoids and M. truncatula, P. sativum and V. faba were chosen to represent IRLC, as shown in Fig. 1I. Fresh plant tissue from each species was incubated in EDTA buffer to induce the condensed confirmation and thus reverse any forisome expansion caused by wounding (Knoblauch et al., 2001). We measured the length (l) and width (w) of individual unstained condensed forisomes in the stem phloem. The parameters showed considerable standard deviations caused by the variable size of the corresponding sieve elements. Therefore, we calculated the more constant average ratio between length and width (r = l/w). Dipteryx panamensis forisomes (r = 8·5) were found to be similar to those from the evolutionarily more recent species C. gladiata (r = 8·1), L. japonicus (r = 8·2), M. truncatula (r = 6·6), P. sativum (r = 8·9) and V. faba (r = 11). We also measured the width of the corresponding sieve plates (spw) to correlate those values with the width (w) of individual forisomes. The correlation coefficient between forisome and sieve plate widths was positive for all species. We obtained values of 0·40 for D. panamensis, 0·63 for C. gladiata, 0·43 for L. japonicus, 0·45 for M. truncatula, 0·43 for P. sativum and 0·24 for V. faba. These data indicate that forisomes are wider in larger sieve elements with wider sieve plates. We also calculated the ratio between sieve plate width and forisome width (v = spw/w) to gain insight into the forisome widening required for sieve element occlusion. In the basal species D. panamensis, this ratio (v = 3·7 ± 0·7) was similar to the values observed in C. gladiata (v = 3·4 ± 0·5), L. japonicus (v = 3·7 ± 0·7), M. truncatula (v = 3·4 ± 1·0), P. sativum (v = 5·6 ± 1·1) and V. faba (v = 4·6 ± 1·0), suggesting that forisome width in each species is likely to be adjusted to achieve efficient sieve element occlusion. However, ANOVA showed that the ratio was slightly elevated in P. sativum and V. faba (P < 0·05).
In silico analysis of group 1 SEO proteins
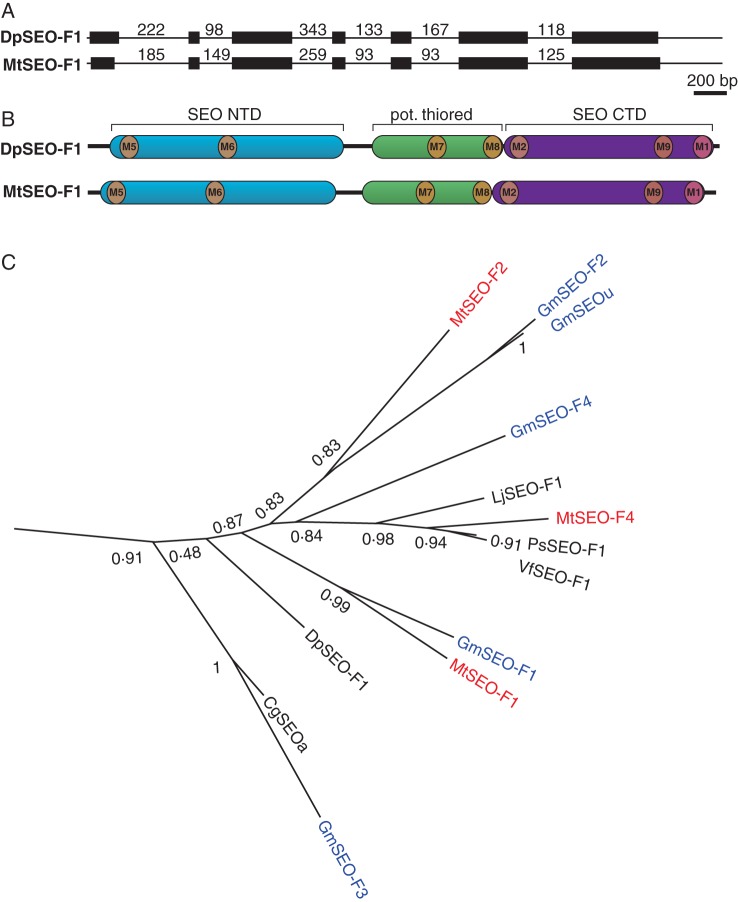
Forisomes are encoded by members of the recently described SEO gene family, which is divided into seven groups, the first three exclusive to Papilionoideae species (Rüping et al., 2010). To gain insight into the evolution of forisomes from a broad spectrum of Papilionoideae species, we amplified a putative SEO-F gene from D. panamensis (DpSEO-F1). The SEO gene family has a conserved exon–intron structure and a unique domain structure comprising SEO-specific N-terminal and C-terminal domains (SEO-NTD, SEO-CTD) flanking a putative thioredoxin fold (Rüping et al., 2010), all of which were present in DpSEO-F1, as shown by alignment with MtSEO-F1 (Fig. 2A, B).
Fig. 2.
Phylogenetic analysis of group 1 SEO proteins. (A) Genomic organization of DpSEO-F1 compared with the common organization of SEO genes, represented by MtSEO-F1. Exons are represented by black boxes and introns by black lines (not to scale, but the length of each intron is indicated in base pairs). (B) Schematic representation of SEO protein domain organization. DpSEO-F1 comprises an N-terminal domain (SEO NTD; blue), a potential thioredoxin fold (pot. thiored, green) and a C-terminal domain (SEO CTD; purple). This specific combination is only present in SEO proteins (Rüping et al., 2010). The positions of previously described motifs (M1, M2; Rüping et al., 2010) and newly identified motifs (M5–M9; Supplementary Data Fig. S2) are indicated. (C) Group 1 of the phylogram created with FastTree2 based on MUSCLE (Edgar, 2004) amino acid alignment (Supplementary Data Fig. S1). SEO proteins previously described by Rüping et al. (2010) as well as DpSEO-F1 and LjSEO-F1 were used to construct the phylogram. The number of amino acid substitutions is related to branch length, and bootstrap values are indicated.
We also identified a putative SEO-F gene in the model legume L. japonicus (LjSEO-F1), allowing us to include the robinoid clade in the phylogenetic analysis. All the SEO proteins previously described by Rüping et al. (2010) were aligned with the novel DpSEO-F1 and LjSEO-F1 sequences. A phylogram was created (Supplementary Data Fig. S1) and group 1 is shown in Fig. 2C. Remarkably, DpSEO-F1 was allocated to group 1, close to the branch containing MtSEO-F1. LjSEO-F1 was also allocated to group 1 and clustered with MtSEO-F4. Because both novel proteins clustered within group 1, we focused on the group 1 SEO proteins.
The amino acid sequences of the group 1 SEO proteins were aligned to gain further evolutionary data (Supplementary Data Fig. S2). Even though the last common ancestor of D. panamensis and the other species existed 58·6 million years ago (Lavin et al., 2005), there was still considerable sequence conservation (63–97 % sequence similarity, e.g. 76·3 % between MtSEO-F1 and DpSEO-F1). The previously identified SEO-specific protein motifs 1 and 2 (Rüping et al., 2010) were more strongly conserved among the group 1 SEO proteins (e.g. 16 of 23 amino acids were identical in motif 1, as shown in Supplementary Data Fig. S2) than among the other groups of the SEO family. The alignment of group 1 SEO proteins identified further regions of strong conservation (M5–M9) featuring at least seven neighbouring amino acids that were identical or shared conserved substitutions (Supplementary Data Fig. S2). Interestingly, these regions were already highly conserved in DpSEO-F1, but differed in the amino acid sequences of P-proteins (e.g. AtSEO and NtSEO; Ernst et al., 2012; Jekat et al., 2013). The phylogenetic data suggest that the sequences of group 1 SEO genes are highly conserved among the Papilionoideae, probably reflecting strong selection pressure on these genes.
Structural analysis of group 1 SEO-F protein complexes
The group 1 SEO proteins MtSEO-F1 and MtSEO-F4 can form homomeric forisome-like protein bodies (Müller et al., 2010), so we investigated whether other group 1 SEO proteins have the same ability. Five potential SEO-F genes (DpSEO-F1, CgSEOa, LjSEO-F1, PsSEO-F1 and VfSEO-F1, corresponding to the native forisomes we investigated) were therefore selected for individual expression in N. benthamiana epidermal cells under the control of the strong, constitutive Cauliflower mosaic virus (CaMV) 35S promoter. The corresponding protein sequences are marked in black in Fig. 2C. We used MtSEO-F1 as a reference.
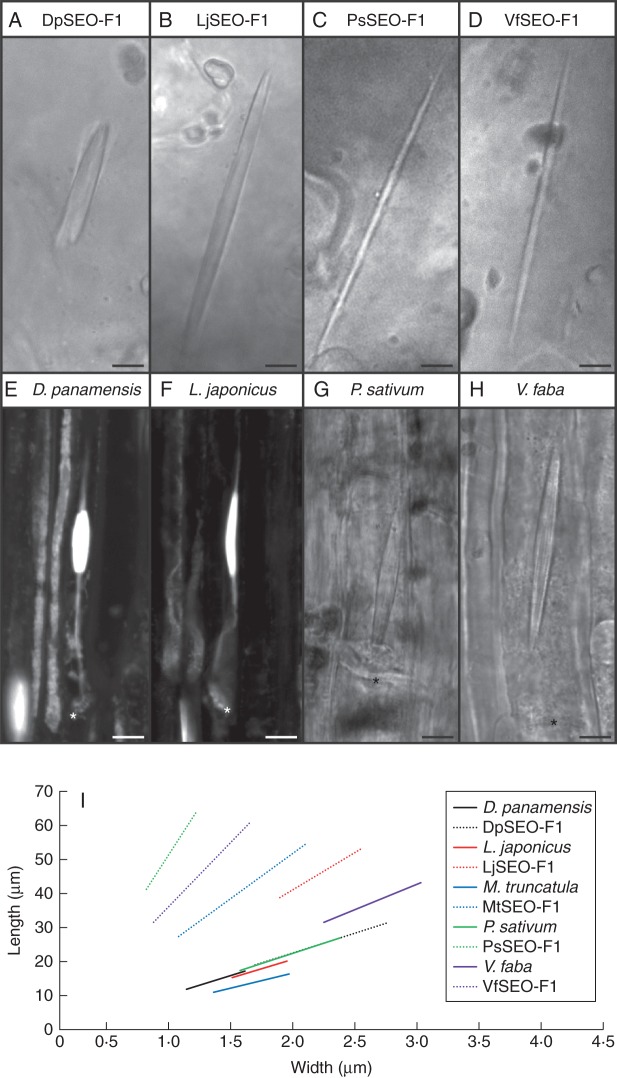
DpSEO-F1 was able to assemble into functional forisome-like structures (Fig. 3A) as also shown by the presence of expanded DpSEO-F1 forisomes, probably resulting from damage caused by the laser. LjSEO-F1 (Fig. 3B), PsSEO-F1 (Fig. 3C) and VfSEO-F1 (Fig. 3D) also assembled into protein bodies with a typical forisome-like spindle shape, similar to the MtSEO-F1 homomeric structure described by Müller et al. (2010), thus confirming they are forisome proteins. Based on the results of the present study the original name of the protein PsSEOa (Rüping et al., 2010) was changed to PsSEO-F1 to reflect the confirmed role in forisome assembly. Interestingly, the DpSEO-F1 and LjSEO-F1 protein bodies did not possess tail-like protrusions, although D. panamensis and L. japonicus produced tailed forisomes (Fig. 3E, F), indicating that forisome tails are not critical for assembly. Exceptionally, CgSEOa did not assemble into forisome-like protein bodies, but a venus-tagged version of CgSEOa was incorporated into MtSEO-F1 and DpSEO-F1 artificial forisomes (Supplementary Data Fig. S3A–C).
Fig. 3.
Morphology of artificial forisomes compared with native forisomes. Artificial forisomes were produced by heterologous expression in N. benthamiana epidermal cells. (A) DpSEO-F1 expression produced short, wide protein bodies. (B) LjSEO-F1 expression produced homomeric forisome-like protein bodies with the typical spindle shape. (C, D) The expression of PsSEO-F1 (C) and VfSEO-F1 (D) produced long, thin protein bodies. (E, F) Native forisomes of D. panamensis (E) and L. japonicus (F) in sieve elements. Sieve plates are marked with an asterisk. Forisomes were stained with sulphorhodamine 101 to visualize the tails. (G, H) Native P. sativum (G) and V. faba (H) forisomes in sieve elements. Sieve plates are marked with an asterisk. (I) Schematic overview summarizing the proportions of artificial and native forisomes (mean length, mean width and mean ratio). The range of the graphs is adjusted to the standard deviations of length and width. Artificial forisomes are represented by dotted lines and corresponding native forisomes are shown by solid lines in the same colour. Scale bars: (A–H) = 5 μm.
We next compared the morphology of homomeric artificial forisomes and native forisomes (Fig. 3A–H). The proportions of DpSEO-F1 recombinant protein bodies were similar to those of D. panamensis native forisomes (Fig. 3E), although they were longer and wider (r = 8·7). Recombinant protein bodies assembled from LjSEO-F1 (r = 16·4), VfSEO-F1 (r = 28·4) and PsSEO-F1 (r = 43·3) were also longer than their native counterparts, but were thinner or comparable in width so the length-to-width ratio was greater than that of the native forisomes in the corresponding species L. japonicus, P. sativum or V. faba (Fig. 3F–H).
The geometrical data were visualized using the graphs shown in Fig. 3I (for standard deviations see Supplementary Data Table S1). The proportions of artificial forisomes varied between different group 1 SEO proteins, whereas the proportions of the native forisomes were similar across species. The geometry of native V. faba forisomes differed slightly and therefore the corresponding graph was steeper. It seemed likely that the variation in the morphology of artificial and native forisomes occurred because more than one SEO-F protein contributes to forisome assembly in most species. Additional SEO-F genes from the sequenced species M. truncatula and Glycine max have previously been identified (Rüping et al., 2010), and therefore we developed a series of experiments focusing on SEO-F genes from the model legume M. truncatula to validate this assumption.
Incorporation of MtSEO-F subunits into native forisomes
Among the four genes that are known to encode forisome proteins in M. truncatula, MtSEO-F1, MtSEO-F2 and MtSEO-F4 belong to group 1, whereas MtSEO-F3 belongs to group 3 (Rüping et al., 2010). Recently, it was shown that MtSEO-F2 does not assemble into homomeric protein bodies, but can be incorporated into artificial forisomes based on MtSEO-F1 or MtSEO-F4 (Müller et al., 2010; Groscurth et al., 2012). In contrast, MtSEO-F3 was not incorporated into MtSEO-F1 or MtSEO-F4 protein bodies expressed in a heterologous system (Supplementary Data Fig. S3D–F; Groscurth et al., 2012). On the other hand, MtSEO-F3 expressed under the control of the MtSEO-F1 promoter was incorporated into native V. faba forisomes in roots (Pélissier et al., 2008). We therefore analysed the behaviour of these proteins in a homologous system (undifferentiated sieve elements in M. truncatula plants) by root transformation with each of the four MtSEO-F genes under the control of native promoters, using C-terminal hrGFP tags for visualization. As a control, hrGFP was expressed using the MtSEO-F1 promoter, revealing cytosolic green fluorescence in undifferentiated sieve elements (clearly recognizable by the presence of vacuoles), confirming that hrGFP does not interact non-specifically with native forisome bodies (Fig. 4A). No fluorescence was observed in mature, translocating sieve elements (data not shown), probably because hrGFP is transported to the root tips and unloaded from the protophloem files as shown for native GFP (Stadler et al., 2005). When hrGFP was fused to MtSEO-F1, MtSEO-F2 or MtSEO-F4 and expressed under the control of the corresponding promoter, we detected fluorescence of the native forisome bodies (Fig. 4B, C, E), whereas tagged MtSEO-F3 was not incorporated into forisomes, but instead formed fluorescent plugs on the sieve plates (Fig. 4D). Because we were unable to confirm the assembly of MtSEO-F3 into homomeric, spindle-shaped protein bodies or the incorporation of MtSEO-F3 into artificial or native forisome bodies, we excluded this group 3 SEO protein from further studies and focused on MtSEO-F1, MtSEO-F2 and MtSEO-F4.
Fig. 4.
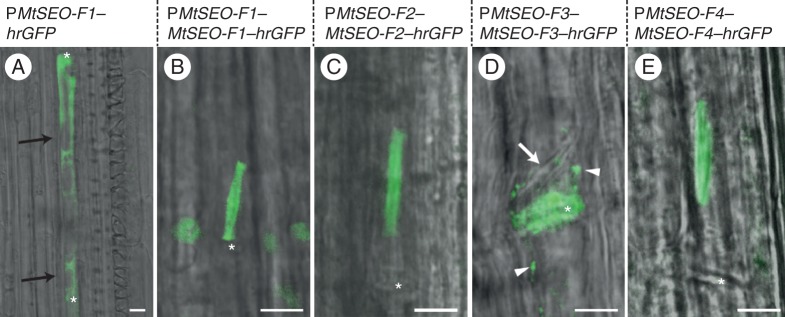
Transformation of M. truncatula roots. The hrgfp control was expressed under the control of the MtSEO-F1 promoter (PMtSEO-F1), whereas fusions of hrgfp to the SEO-F genes MtSEO-F1, MtSEO-F2, MtSEO-F3 or MtSEO-F4 were expressed under the control of the promoter of the corresponding SEO-F gene (PMtSEO-F1, -F2, -F3 or -F4). (A) PMtSEO-F1–hrGFP was expressed as a control and green fluorescence was detected in young sieve elements, identified by the presence of vacuoles (black arrows). (B, C, E) Expression of PMtSEO-F1-MtSEO-F1–hrGFP((B), PMtSEO-F2-MtSEO-F2–hrGFP((C) and PMtSEO-F4-MtSEO-F4–hrGFP (E) produced fluorescent forisomes due to the incorporation of the MtSEO-F subunit into native forisomes. (D) Expression of PMtSEO-F3-MtSEO-F3–hrGFP produced fluorescent agglomerates clearly visible on sieve plates (asterisk), but they were not incorporated into native forisomes (white arrow). Furthermore, parietal green fluorescence was detected in the sieve elements (arrowheads). Sieve plates are marked with an asterisk. Scale bars: (A–E) = 5 μm.
Co-expression of MtSEO-F1, MtSEO-F2 and MtSEO-F4
Artificial forisomes encoded by MtSEO-F1 have different geometrical parameters compared with native M. truncatula forisomes, indicating that forisome assembly is dependent on several SEO-F subunits in this species. Therefore, we co-expressed all possible combinations of MtSEO-F1, MtSEO-F2 and MtSEO-F4 in N. benthamiana epidermal cells and investigated the geometrical parameters, ultrastructure and functionality of the resulting complexes. As references, homomeric MtSEO-F1 and MtSEO-F4 artificial forisomes were produced under the same conditions (Müller et al., 2010).
Morphology and reactions
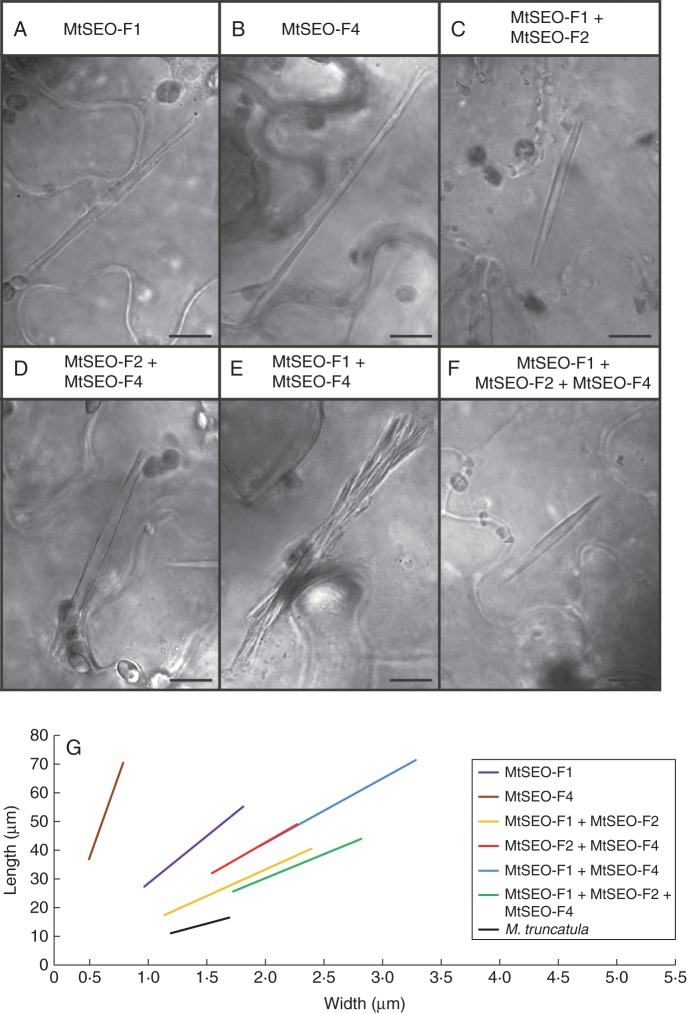
The co-expression of two MtSEO-F subunits produced heteromeric protein bodies with a different length-to-width ratio compared with homomeric MtSEO-F1 (r = 20; Fig. 5A) or MtSEO-F4 (r = 70; Fig. 5B). The co-expression of MtSEO-F2 with MtSEO-F1 (r = 11; Fig. 5C) or MtSEO-F4 (r = 14; Fig. 5D) therefore generated compact protein bodies that were more densely packed than the corresponding homomeric complexes and resembled native forisomes more closely. In contrast, the co-expression of MtSEO-F1 and MtSEO-F4 resulted in protein bodies featuring loosely attached fibre bundles that did not come together at the tips of the structure (Fig. 5E). However, single fibre bundles maintained the typical spindle shape. Their size was approximately 30 % of that of the entire recombinant protein body (average l = 14·3 μm; average w = 0·9 μm) and their ratio (r = 15) was similar to that of MtSEO-F1 + MtSEO-F4 protein bodies (r = 14).
Fig. 5.
Structural aspects of the three known M. truncatula forisome subunits expressed in N. benthamiana epidermal cells. Morphology is clearly dependent on the subunits. (A, B) MtSEO-F1 (A) and MtSEO-F4 (B) artificial forisomes are long and thin. (C, D) MtSEO-F1 + MtSEO-F2 (C) and MtSEO-F2 + MtSEO-F4 (D) protein bodies show the typical spindle shape with dense packing. (E) MtSEO-F1 + MtSEO-F4 artificial forisomes comprise attached fibres that do not form a typical compact protein body. (F) Combinatorial expression of subunits MtSEO-F1, MtSEO-F2 and MtSEO-F4 produced artificial forisomes with dense packing and a smooth surface, morphologically similar to native forisomes. (G) Schematic overview showing the proportions of artificial forisomes containing MtSEO-F proteins compared with native M. truncatula forisomes. The measured geometrical data (mean length, mean width and mean ratio) are visualized in the corresponding linear graphs. The range of the graphs is adjusted to the standard deviations of length and width. Homomeric protein bodies (purple and brown line) displayed the highest length-to-width ratio, whereas the combinatorial expression of subunits generated shorter and wider protein bodies (red, blue and yellow lines). The size of MtSEO-F1 + MtSEO-F2 + MtSEO-F4 protein bodies (green line) was closest to that of native forisomes (black line). Scale bars: (A–F) = 10 μm.
The sequence similarities among the three genes made it difficult to produce expression cassettes containing all three MtSEO-F sequences, so the combination of all three subunits was co-expressed by co-infiltrating N. benthamiana cells with two pBIN19 plasmids (see Materials and methods for further details). The morphology of the resulting protein bodies (r = 10) was shifted even further towards the parameters measured in native forisomes and the surface of the protein bodies seemed smooth and tightly packed (Fig. 5F). The geometric data for each combination of subunits is summarized in Table 1 and illustrated together with values for native M. truncatula forisomes in Fig. 5G. This demonstrates that the morphology of protein bodies generated by the combinatorial expression of MtSEO-F1, MtSEO-F2 and MtSEO-F4 is closest to that of native forisomes (r = 7).
Table 1.
Proportions and Ca2+-dependent responses of native M. truncatula forisomes and artificial forisomes comprising MtSEO-F subunits
| Protein body | Length (μm) | Width (μm) | Ratio | Volume increase |
|---|---|---|---|---|
| MtSEO-F1 | 40 ± 10 | 2·0 ± 0·7 | 20·0 | × 5–15 (Noll et al., 2011) |
| MtSEO-F4 | 50 ± 10 | 0·7 ± 0·2 | 69·8 | × 7 (Noll et al., 2011) |
| MtSEO-F1 + MtSEO-F2 | 30 ± 10 | 2·6 ± 0·9 | 11·2 | × 1·5–2·5 |
| MtSEO-F2 + MtSEO-F4 | 40 ± 8 | 2·9 ± 0·5 | 14·1 | × 1·7–3·5 |
| MtSEO-F1 + MtSEO-F4 | 50 ± 10 | 4·1 ± 1·0 | 13·7 | × 1·9–5 |
| MtSEO-F1 + MtSEO-F2 + MtSEO-F4 | 35 ± 9 | 3·5 ± 0·7 | 10·1 | Not examined |
| M. truncatula wild-type | 14 ± 3 | 2·1 ± 0·3 | 6·8 | × 2·1–5·2 |
Lengths and widths are given as mean ± s.d.
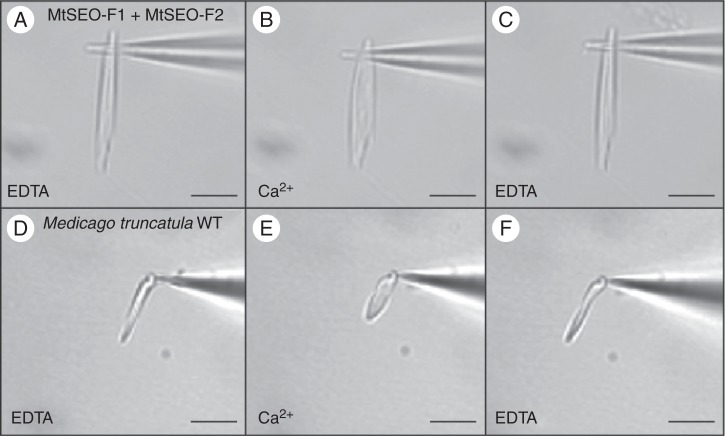
The morphology of the condensed forisomes clearly depended on the presence of particular subunits, so we next focused on the influence of different subunits on the extent of the conformational change. Artificial forisomes were isolated and attached to glass needles. Their expansion was then triggered by the application of 100 µm Ca2+. Protein bodies representing all examined subunit combinations underwent rapid conformational changes that were fully reversible, as shown for MtSEO-F1 + MtSEO-F2 protein bodies in Fig. 6A–C. We evaluated five independent protein bodies representing each combination and calculated the volume increase (Table 1). Unfortunately, we could not purify artificial forisomes comprising all three subunits because the two-vector approach makes it impossible to determine whether isolated artificial forisomes represent two or three MtSEO-F proteins (the cytoplasm-localized fluorescent reporter is lost when the cells are disrupted). Homomeric artificial forisomes showed both a high volume increase and a high variance (Noll et al., 2011). In contrast, combinatorial expression resulted in protein bodies with lower and more constant values (1·5- to 5-fold; Table 1). Native M. truncatula forisomes were isolated and exposed to the same experimental conditions (Fig. 6D–F), resulting in a 2- to 5-fold volume increase, similar to the 3- to 5-fold increase described for V. faba (Schwan et al., 2009). These data showed that native and heteromeric artificial forisomes undergo a similar volume increase during expansion, whereas homomeric artificial forisomes swell much more (Noll et al., 2011).
Fig. 6.
Reversible conformational change of forisomes ex vivo. (A–C) Artificial MtSEO-F1 + MtSEO-F2 forisomes. (D–F) Native M. truncatula forisomes. Isolated forisomes were attached to a glass needle (A, D) and expansion was triggered by applying 100 μm Ca2+ (B, E). The reaction was reversed by applying EDTA buffer (C, F). Scale bars = 10 μm.
Ultrastructure
We next analysed the ultrastructure of homomeric and heteromeric artificial forisomes by electron microscopy to determine the basis of the observed differences in morphology and behaviour. SEM analysis showed that MtSEO-F4, MtSEO-F1 + MtSEO-F4, MtSEO-F1 + MtSEO-F2 and MtSEO-F2 + MtSEO-F4 protein bodies always comprised several densely packed cuboid fibres, similar to those described for MtSEO-F1 (Groscurth et al., 2012). Structural differences observed by light microscopy were also confirmed: MtSEO-F4 artificial forisomes (Fig. 7A) were thinner than MtSEO-F1 + MtSEO-F2 (Fig. 7B) and MtSEO-F2 + MtSEO-F4 (Fig. 7C) protein bodies. SEM images also showed that MtSEO-F1 + MtSEO-F4 fibrils did not assemble into a uniform protein body (Fig. 7D). Furthermore, higher magnifications confirmed that co-expression produced protein bodies with smoother surfaces compared with homomeric complexes; e.g. compare homomeric MtSEO-F4 (Fig. 7E) with MtSEO-F1 + MtSEO-F2 (Fig. 7F) and MtSEO-F2 + MtSEO-F4 (Fig. 7G). The separation of fibrils in the MtSEO-F1 + MtSEO-F4 protein bodies was clearly detected (Fig. 7H).
Fig. 7.
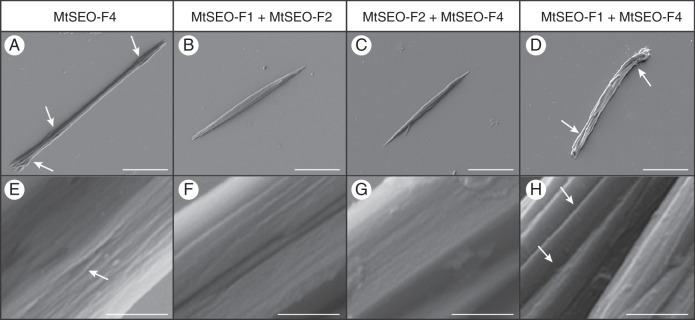
Scanning electron micrographs of artificial forisomes expressed in N. benthamiana leaves. (A) MtSEO-F4 protein bodies are thin in the centre and appear more rod-like than spindle-shaped, but single-fibril bundles can still be discerned at the tips (arrows). (B, C) MtSEO-F1 + MtSEO-F2 (B) and MtSEO-F2 + MtSEO-F4 (C) artificial forisomes possess the typical spindle shape. (D) MtSEO-F1 + MtSEO-F4 protein bodies comprise loosely attached fibres with distinct inter-fibre spaces (arrows). (E) Higher magnification of MtSEO-F4 reveals shallow cavities (arrow) in the protein body. (F, G) The surface of MtSEO-F1 + MtSEO-F2 (F) and MtSEO-F2 + MtSEO-F4 (G) artificial forisomes is tightly packed. (H) Magnification of an MtSEO-F1 + MtSEO-F4 protein body shows distinct fibres with large intervening cavities (arrows). Scale bars: (A–D) = 10 μm, (E–H) = 500 nm.
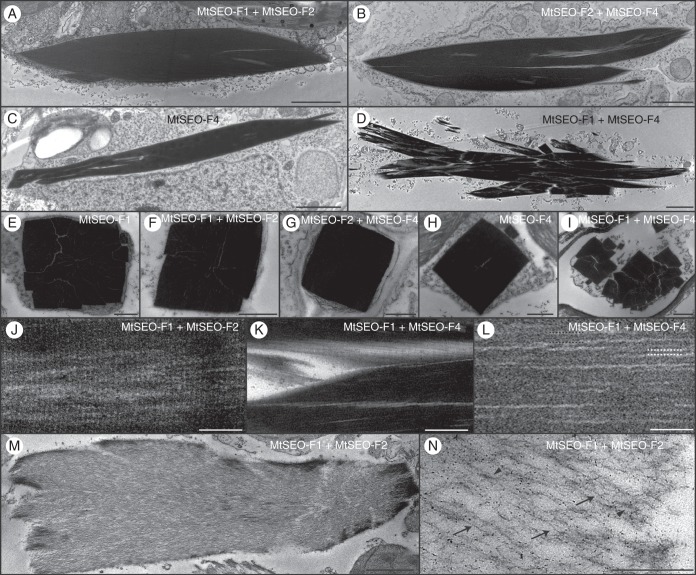
The TEM images of longitudinal sections from MtSEO-F1 + MtSEO-F2 (Fig. 8A) and MtSEO-F2 + MtSEO-F4 (Fig. 8B) protein bodies showed that the proportions of artificial forisomes were similar to those of native forisomes, recognizable by the spindle shape becoming wider in the centre of the recombinant protein body. The protein bodies were also electron-dense, suggesting tightly packed fibres, and single fibril bundles could be identified only occasionally. Homomeric MtSEO-F4 forisomes were thinner and less densely packed, but spaces were visible between fibres and the outline was less uniform (Fig. 8C). Fibres in the MtSEO-F1 + MtSEO-F4 forisomes were densely packed, but did not assemble into a single dense recombinant protein body, leading to several clear spaces between the fibres (Fig. 8D). Cross-sections of condensed forisomes representing all combinations of MtSEO-F proteins reflected data that were obtained previously by CLSM and SEM in the present article (Fig. 8E–I); e.g. MtSEO-F1 + MtSEO-F4 fibres were not united into one compact protein body (Fig. 8I). Fine cross-striations, like those present in native forisomes, were identified in the longitudinal sections of all combinations (shown for MtSEO-F1 + MtSEO-F2 as an example in Fig. 8J). The periodicity of cross-striations ranged from 11 to 13·5 nm in all the artificial forisomes (Table 2). These data show that the ultrastructural patterns beneath the level of fibre packing remain almost the same, regardless of the subunits involved.
Fig. 8.
Representative transmission electron micrographs of artificial forisomes expressed in N. benthamiana leaves. (A–D) Longitudinal sections of condensed protein bodies following the combinatorial expression of MtSEO-F subunits. MtSEO-F1 + MtSEO-F2 (A) and MtSEO-F2 + MtSEO-F4 (B) protein bodies are densely packed, whereas MtSEO-F4 protein bodies (C) reveal some spaces. MtSEO-F1 + MtSEO-F4 protein bodies (D) reveal even more spaces between dense fibril bundles. (E–I) Cross-sections of condensed protein bodies following the expression of MtSEO-F1 (E), MtSEO-F1 + MtSEO-F2 (F), MtSEO-F2 + MtSEO-F4 (G), MtSEO-F4 (H) and MtSEO-F1 + MtSEO-F4 (I). With the exception of (I), all protein bodies were characterized by a cuboid outline and dense packing. In MtSEO-F1 + MtSEO-F4 protein bodies (I), the fibre bundles were separated from each other. (J) Higher magnification of a longitudinal section of the MtSEO-F1 + MtSEO-F2 protein body revealed a fine cross-striation pattern perpendicular to the forisome body. (K, L) Higher magnification of a longitudinal section of the MtSEO-F1 + MtSEO-F4 protein body. (K) Coarse cross-striation. (L) Fibrils (black dotted lines) and filaments (white dotted lines). (M, N) Expanded MtSEO-F1 + MtSEO-F2 protein body following the application of Ca2+ during embedding. (M) The outline of the protein body remains condensed, indicating that the expansion proceeds from the central body towards the periphery. (N) Single filaments (arrows) and fibrils (arrowheads) are distinguishable. Scale bars: (A–D, M) 2 μm, (E–I, K, N) 500 nm, (J, L) 100 nm.
Table 2.
Periodicity of cross-striations in artificial forisomes
| Protein body | Fine cross-striation (nm) | Coarse cross-striation (nm) |
|---|---|---|
| MtSEO-F1 | 12·5 ± 1·2 | None |
| MtSEO-F4 | 11·0 ± 0·6 | 42·2 ± 6 |
| MtSEO-F1 + MtSEO-F2 | 13·2 ± 0·9 | None |
| MtSEO-F2 + MtSEO-F4 | 11·6 ± 1·6 | 50·5 ± 2·5 |
| MtSEO-F1 + MtSEO-F4 | 13·3 ± 0·6 | 47·5 ± 4·6 |
Data are mean ± s.d.
Additional coarse cross-striations with a periodicity of 42–50 nm were observed in the MtSEO-F4, MtSEO-F1 + MtSEO-F4 and MtSEO-F2 + MtSEO-F4 protein bodies (shown for MtSEO-F1 + MtSEO-F4 as an example in Fig. 8 K; Table 2) but not in the homomeric MtSEO-F1 or heteromeric MtSEO-F1 + MtSEO-F2 artificial forisomes. Such striation has only been reported in forisome tails and further investigations are needed to interpret this pattern in tailless artificial forisomes comprising M. truncatula SEO-F proteins. In all protein bodies, filament width ranged from 3 to 5 nm (Fig. 8L), which is similar to the 3–4 nm observed in native M. truncatula forisomes. Furthermore, fibrils with a diameter of 8–15 nm were observed in all combinations, but the dense packing made these structures difficult to discern in the condensed state. The application of Ca2 + before fixation led to the expansion of all artificial forisomes and the morphology was similar to that of expanded native forisomes, as shown for the MtSEO-F1 + MtSEO-F2 forisome as an example (Fig. 8M). Forisome expansion allowed the observation of fibril bundles, single fibrils and even filaments (Fig. 8N), and the size of the filaments in all combinations was in the range 4–6 nm, similar to the 4–5·5 nm filaments observed in expanded M. truncatula forisomes. These ultrastructural parameters at the sub-fibre level were similar for different subunit combinations, in contrast to the structural differences that were observed by light microscopy, with the exception of the coarse cross-striation that was only observed in the presence of MtSEO-F4.
Expression of MtSEO-F2 with SEO-F genes from different species
Finally we investigated the apparent critical role of MtSEO-F2 in the formation of correctly proportioned M. truncatula forisomes by co-expressing MtSEO-F2 with SEO-F genes from other species, namely DpSEO-F1, LjSEO-F1, PsSEO-F1 and VfSEO-F1. The geometrical parameters of all the hybrid forisomes were modified without affecting the typical spindle shape (Fig. 9). The hybrid protein bodies were fatter than the homomeric structures, thus shifting towards the proportions of the native forisomes in each species, i.e. LjSEO-F1 + MtSEO-F2 (r = 12), PsSEO-F1 + MtSEO-F2 (r = 15) and VfSEO-F1 + MtSEO-F2 (r = 11), which was the same ratio as that observed for native V. faba forisomes. DpSEO-F1 + MtSEO-F2 protein bodies were similar to homomeric DpSEO-F1 protein bodies and the ratio was maintained (r = 8).
Fig. 9.
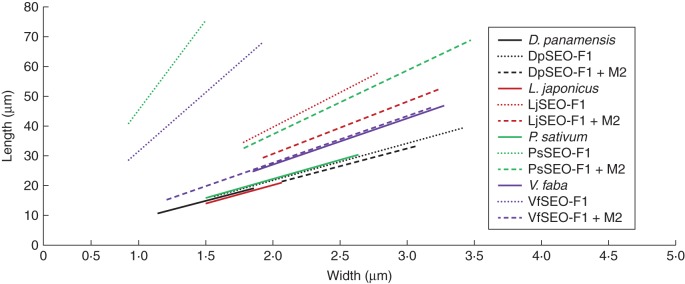
Schematic overview summarizing the size of heteromeric protein bodies after co-expression of different SEO-F genes with MtSEO-F2, compared with homomeric protein bodies and native forisomes. MtSEO-F2 is abbreviated to M2 in the key. The values are visualized in the corresponding linear graphs. Homomeric artificial forisomes are represented by dotted lines, heteromeric artificial forisomes by dashed lines and native forisomes by solid lines. Mean length, mean width and mean ratio were calculated and used to draw the corresponding linear graphs, whereas the range of the graph is determined by standard deviations of length and width. Corresponding protein bodies are displayed in the same colour (see key). Most artificial forisomes increased in width when MtSEO-F2 was co-expressed, resulting in flatter graphs, with DpSEO-F1 + MtSEO-F2 as the exception. The slope of the VfSEO-F1 + MtSEO-F2 graph was similar to that of the graph for native V. faba forisomes.
DISCUSSION
Forisome structure is evolutionarily conserved
Previous TEM studies concerning forisomes have focused mainly on species representing the milletioids, robinoids and IRLC (Palevitz and Newcomb, 1971; Esau, 1978; Lawton, 1978a, b). Prior to this investigation, there were no molecular data and only limited structural data available for first-branching papilionoid lineages, the latter including the detection of expanded forisomes in Swartzia spp. by light microscopy (Behnke, 1981; Peters et al., 2010). To broaden our molecular and structural analysis of forisomes in the Papilionoideae, in the current study we included D. panamensis, a species of the tribe Dipterygeae (an early branch of the papilionoid lineage) whose forisomes have not been investigated thus far (Cardoso et al., 2012). Bright-field microscopy confirmed that forisomes with the typical spindle-shaped morphology are present in D. panamensis and can undergo a conformational change that probably allows them to control the mass flow of photoassimilates. These forisomes were compared with those from the Old World clade, i.e. the species C. gladiata, L. japonicus, M. truncatula, P. sativum and V. faba (Lavin et al., 2005). In all species, we observed a positive correlation between forisome width and that of the corresponding sieve element, indicating that larger sieve elements contain larger forisomes. This also explains the significant variances in the length and width of forisomes from each species. The proportions of forisomes (length-to-width ratio) are evolutionarily conserved and forisome width is 3·4- to 5·6-fold smaller than that of the corresponding sieve plate. However, statistical analysis showed that the ratio between forisome width and sieve plate width is slightly greater in P. sativum and V. faba. Additional studies are needed to determine whether this increase corresponds to further forisome widening during the conformational change in these species and whether changes in the SEO-F subunit composition and/or the amino acid sequences of the forisome subunits mediate this difference.
The ultrastructures of forisomes from D. panamensis and the Old World clade were found to be similar, including fibril size and the periodicity of cross-striations (Zee, 1969; Wergin and Newcomb, 1970; Palevitz and Newcomb, 1971; Wergin et al., 1975; Lawton, 1978b; Arsanto, 1982). The conservation of forisome morphology, ultrastructure and function since the speciation of D. panamensis indicates that SEO-F proteins have been under strong selection pressure since they diverged from conventional P-proteins, probably by gene duplication and subsequent neofunctionalization. The basis of the selection pressure is unknown, although it may reflect the reduction in fitness caused by photoassimilate loss and/or pathogen susceptibility after wounding, even when additional occlusion mechanisms are present. In M. truncatula, occlusion can be mediated by forisomes, by conventional P-proteins encoded by homologous SEO genes (MtSEOa, MtSEOb, MtSEOc and MtSEOe) and by callose synthesis after wounding. This suggests that forisomes significantly improve the sensitivity, efficiency or response time of the occlusion reaction in comparison with the other mechanisms and/or that the reversibility of the forisome reaction is highly beneficial to the plants.
Loss-of-function mutants are not available, and would be difficult to produce because of the multiple (often clustered) SEO-F genes in many species (Rüping et al., 2010). Recently, SEO-specific RNA interference (RNAi) lines were created to prevent the formation of P-proteins in Nicotiana tabacum and Arabidopsis thaliana (Ernst et al., 2012; Jekat et al., 2013). A similar approach in Papilionoideae could be used to dissect the functions of forisomes in detail, as well as their functional interactions with conventional P-proteins. The basal species D. panamensis and late-branching species such as M. truncatula contain both forisomes and conventional P-proteins in the same sieve tubes, so it would be interesting to test RNAi lines lacking either forisomes or conventional P-proteins to assess their relative impact on photoassimilate loss and the evolutionary advantage of having both proteins in the same sieve tubes. Forisomes and P-proteins may respond to different triggers or stimulus intensities. Forisomes expand in response to distant mechanical injury, burning or laser irradiation via the propagation of electropotential waves (Knoblauch and van Bel, 1998; Furch et al., 2007). P-proteins also respond to mechanical injury and laser irradiation (Knoblauch and van Bel, 1998), but the precise trigger is unknown and their dependence on Ca2+ is debated (Will and van Bel, 2006). In addition to forisomes and P-proteins, callose synthesis represents a slow wound reaction and disrupting P-plastids are thought to occlude sieve elements after mechanical injury (King and Zeevaart, 1974; Knoblauch and van Bel, 1998; Furch et al., 2007). The concurrence of different plugging mechanisms makes it difficult to determine their individual roles and evolutionary advantages more precisely.
D. panamensis (Dipterygeae) forisomes include tail-like protrusions, but the function and evolution of these tails is poorly characterized. Certain neighbouring tribes possess tailed (Swartzoideae) or tailless (Brongniartieae) forisomes, indicating that the occurrence of tails does not follow a strict evolutionary pattern (Peters et al., 2010). This may reflect the evolutionary loss of the corresponding genes or the tendency for tail genes to be switched off in some species (Peters et al., 2010). We assume that SEO-F genes are responsible for forisome tails because, as shown (Fig. 1G, H), the ultrastructural fine striations of tails are similar to those of the condensed main forisome bodies (Wergin and Newcomb, 1970; Lawton, 1978b). The D. panamensis genome may contain SEO-F genes required for tail formation, but there are no SEO-specific sequence data available for this species. Interestingly, artificial D. panamensis and L. japonicus forisomes produced by the expression of single SEO-F genes were shown to be tailless, demonstrating that tails are not necessary for the assembly of the main forisome body.
Gene duplications could facilitate the specialization of SEO-F proteins
We extended our earlier phylogenetic analysis of SEO proteins (Rüping et al., 2010) to include novel SEO-F proteins from evolutionarily more distant species (Fig. 2C). In addition to the previously described SEO-specific motifs (Rüping et al., 2010), further conserved sequences were identified (M5–M9) differing significantly from the corresponding sequences of conventional P-proteins (e.g. AtSEO and NtSEO). The motifs M5 and M8 appear to be particularly relevant in group 1 proteins (Supplementary Data Fig. S2). The neofunctionalization of forisome genes may therefore reflect the accumulation of mutations in the conserved regions M5–M9, which possibly converted them from amorphous P-proteins capable of wound-induced irreversible plugging (Knoblauch and van Bel, 1998) to spindle-shaped forisomes with the ability to undergo reversible Ca2+-induced contraction (Knoblauch et al., 2003). In future investigations, we will use site-directed mutagenesis in regions M5–M9 to identify amino acids that are crucial for forisome structure and activity.
The combination of experimental and phylogenetic analysis revealed that the ability of group 1 SEO-F proteins to assemble into macromolecular protein spindles reflects the strong conservation of their amino acid sequences. The proportions of native and artificial forisomes from D. panamensis and protein bodies composed of DpSEO-F1 + MtSEO-F2 (Figs 3A and 9) indicate that the main body of native D. panamensis forisomes is solely composed of DpSEO-F1. In contrast, artificial forisomes from evolutionarily more derived species were generally longer and slimmer than those from their native counterparts (Fig. 3I). Medicago truncatula forisomes assemble from multiple subunits, encoded by SEO-F genes that have probably arisen by duplication after the divergence of D. panamensis and M. truncatula (Lavin et al., 2005; Rüping et al., 2010). The presence of several SEO-F genes in M. truncatula and G. max (Rüping et al., 2010) suggests that additional SEO-F genes may also be present in L. japonicus, P. sativum and V. faba. The co-expression of MtSEO-F2 with artificial forisomes from these species revealed that the morphology of VfSEO-F1, PsSEO-F1 and LjSEO-F1 protein bodies was shifted towards the size of their corresponding native forisomes by the presence of MtSEO-F2 (Fig. 9).
The presence of MtSEO-F1, MtSEO-F2 and MtSEO-F4 on chromosome 1 indicates that these genes did not arise during the early palaeoduplication event described for several Papilionoideae clades (Pfeil et al., 2005; Bertioli et al., 2009) but in subsequent tandem duplication events that most likely predated the divergence of M. truncatula and G. max at least 54 million years ago (Lavin et al., 2005; Rüping et al., 2010). Duplicated genes are only maintained if they have a neutral or positive impact on the species, e.g. due to a positive gene dosage effect. Because the subunits have different effects on forisome assembly (Fig. 5), neofunctionalization or subfunctionalization must have taken place following duplication, apparently resulting in proteins that promote forisome elongation (MtSEO-F1 and MtSEO-F4) or widening (MtSEO-F2).
Species with heteromeric forisomes may therefore benefit through their ability to fine-tune forisome size and activity, providing opportunities to adjust during exposure to different forms of biotic and abiotic stress. The presence of three structural MtSEO-F subunits in M. truncatula may allow more precise tuning of morphological and functional properties, whereas MtSEO-F2 cannot form homomeric forisome-like structures but instead controls the dense packing of the protein body. In contrast, the main body of D. panamensis forisomes may contain only one subunit that assembles in the appropriate forisome geometry. However, the presence of additional SEO-F proteins in D. panamensis cannot be ruled out due to the lack of comprehensive genomic and cDNA data.
Role of different forisome subunits in M. truncatula
In previous studies (Rüping et al., 2010), nine SEO genes were identified in the M. truncatula genome and further analysis showed that MtSEO-F1, MtSEO-F2 and MtSEO-F4 represent group 1 SEO proteins and are involved in forisome assembly (Fig. 4B, D, E; Müller et al., 2010; Groscurth et al., 2012). Further MtSEO proteins (MtSEOa to MtSEOe) clustered in group 4, but this group includes SEO proteins from non-Fabaceae species, probably assembling to conventional P-proteins, and was therefore largely excluded from the current study (Rüping et al., 2010; Ernst et al., 2012). Exceptionally, MtSEO-F3 is found in the Fabaceae-specific group 3 of the SEO protein family and the function of this protein has been discussed (Pélissier et al., 2008; Müller et al., 2010; Rüping et al., 2010; Groscurth et al., 2012; Supplementary Data Fig. 3D–F). Here we found that the location of an MtSEO-F3–hrGFP fusion protein was similar to that of fluorescence-tagged NtSEO and AtSEO proteins from groups 5 and 6 (Fig. 4D; Ernst et al., 2012). Peléssier et al. (2008) reported the incorporation of MtSEO-F3-GFP into V. faba forisomes, thus conflicting with our results, but this could be attributed to the different type of GFP (conventional GFP instead of hrGFP), the different expression host (V. faba instead of M. truncatula) or the different promoter (PMtSEO-F1 instead of PMtSEO-F3). The different forms of GFP have similar molecular weights (GFP 26·9 kDa, hrGFP 27·3 kDa), but the conformation of hrGFP could hinder incorporation whereas conventional GFP could be more permissive. Alternatively, distinct forms of protein packing in M. truncatula and V. faba forisome bodies may support the incorporation of GFP fusion proteins in the latter but not in the former. The different promoters could potentially become active at different time points during late sieve element differentiation, and thus the corresponding promoter should be used in localization studies to ensure accuracy. We were unable to determine the precise reason for the differences between the studies, so the role of the reporter protein, expression host and promoter need to be addressed in more detail.
The phylogenetic analysis suggested that group 3 SEO proteins such as MtSEO-F3 may represent an intermediate form during the evolution of conventional P-proteins (group 5) into forisomes (group 1) (Rüping et al., 2010). MtSEO-F3 may possess characteristics common to both conventional P-proteins and forisomes, and may potentially interact with forisomes (Supplementary Data Fig. S3E, F). Indeed, mass spectrometry analysis of purified M. truncatula forisomes identified MtSEO-F3 peptide sequences (Rüping et al., 2010). These could represent co-purification artefacts, given that MtSEO-F3 is probably located close to forisomes in the sieve element, and because all four proteins have a similar molecular weight they will subsequently all be found in the 75-kDa protein band used for sequencing. We therefore excluded MtSEO-F3 from our investigation and investigated the influence of MtSEO-F1, MtSEO-F2 and MtSEO-F4 on artificial forisome assembly by CLSM, SEM and TEM.
Comparisons of homomeric and heteromeric artificial forisomes revealed differences in morphology and reactivity, depending on the subunit combination (Fig. 5A–F; Table 1). The quantitative characterization of heteromeric artificial forisomes showed that MtSEO-F2 (Figs 7B, C and 8A, B) appears to promote inter-fibril interactions but has a lower capacity for longitudinal polymerization, which must reflect the accumulation of specific mutations during the divergence of the MtSEO-F proteins. The precise localization of the MtSEO-F2 protein, either within or between the MtSEO-F1/MtSEO-F4 filaments, remains to be determined. The uniform size of single MtSEO-F1 + MtSEO-F4 fibres indicated that these protein bodies are not assembled from homomeric MtSEO-F1 or MtSEO-F4 fibres, but must represent the heteromeric assembly of both subunits. However, the subsequent assembly of MtSEO-F1 + MtSEO-F4 fibres into the spindle-shaped protein body is disrupted, probably due to weak or blocked fibre–fibre interaction sites, resulting in loosely packed protein bodies (Figs 5E and 7D, H). As expected, the co-assembly of all three MtSEO-F subunits resulted in protein bodies with proportions that were the most similar to those of native forisomes (Fig. 5F).
A TEM analysis indicated there were no substantial differences in filament width or fine cross-striation patterns between homomeric and heteromeric protein bodies and native forisomes (Arsanto, 1982). The resolution of TEM and the dense packing of the forisome bodies may hide minor differences, which should be investigated using alternative techniques, such as high-resolution TEM, fibre diffraction, nanodiffraction or electron crystallography (Quintana et al., 2004; McDonald et al., 2012; Fujiyoshi, 2013). Surprisingly, artificial forisomes based on MtSEO-F4, e.g. MtSEO-F2 + MtSEO-F4, showed additional coarse cross-striations that have previously been observed solely in forisome tails and may reflect higher orders of structural organization (Fig. 8 K).
The Ca2+-induced expansion tests indicated that isolated heteromeric artificial forisomes swelled less than homomeric protein bodies (Table 1). This may reflect the conservation of strong fibril–fibril interactions in the heteromeric protein bodies during expansion, e.g. pronounced lateral MtSEO-F2 connections, but the ability of calcium ions to access the interaction domains could also be influenced by the presence of additional SEO-F subunits. The filaments, fibrils and fibres of homomeric and heteromeric protein bodies must be connected by strong protein–protein interactions, possibly involving disulphide bonds, otherwise the protein bodies would fall apart during expansion and would not be able to re-establish forisome-like protein bodies during condensation. The orientation of protein domains and the configuration and juxtaposition of disulphide bonds are likely to differ between homomeric and heteromeric complexes, thus influencing their reactivity. The conformational change of heteromeric protein bodies was more similar to the behaviour of native forisomes, but it is necessary to consider the artificial nature of the assay, including the non-phloem biochemical background and the use of a strong constitutive promoter for transgene expression. The formation of accurate native-like forisomes is probably achievable only if the subunits are expressed at the correct physiological levels. Nevertheless, the characterization of the different forisome subunits provided insight into the specific roles of these proteins in forisome assembly and activity, and the potential evolutionary benefits of homomeric and heteromeric native forisomes to optimize the process of sieve element occlusion.
In conclusion, our data provide novel insight into the structural and molecular conservation of group 1 SEO-F proteins. The structural differences between native forisomes and homomeric protein bodies (comprising a single SEO-F subunit) expressed in a heterologous background suggested that combinations of SEO-F proteins may fine-tune the geometric proportions of forisomes and hence their reactivity. This hypothesis was confirmed by the combinatorial expression of M. truncatula SEO-F proteins, producing a range of protein bodies with subtle differences in structure and geometry. This process of fine-tuning was demonstrated explicitly by introducing MtSEO-F2 into homomeric protein bodies comprising single SEO-F subunits from different species. The presence of the heterologous subunit resulted in pronounced lateral forisome growth, suggesting that similar fine-tuning mechanisms occur in the forisomes of all papilionoid legumes.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We gratefully acknowledge Cynthia Barboza Aguilar and Ethel Sánchez Chacón (CIEMIC, San José, Costa Rica) for help with the analysis of D. panamensis forisomes, and Till Matzat and Felix Barbatz (University of Münster) for transmission electron microscopy support. We thank Jan Oltmanns and Bernhard Blob for assistance with root transformation. This research was partially funded by grants from the Fraunhofer Society and the Europäischer Fonds für regionale Entwicklung – Investition in unsere Zukunft (FKZ 300263702).
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson R, Cronshaw J. Sieve-plate pores in tobacco and bean. Planta. 1970;91:173–180. doi: 10.1007/BF00385475. [DOI] [PubMed] [Google Scholar]
- Arsanto JP. Observations on P-protein in dicotyledons. Substructural and developmental features. American Journal of Botany. 1982;69:1200–1212. [Google Scholar]
- Behnke HD. Swartzia: phloem ultrastructure supporting its inclusion into Leguminosae-Papilionideae. Iselya. 1981;2:13–16. [Google Scholar]
- Behnke HD. Nondispersive protein bodies in sieve elements: a survey and review of their origin, distribution and taxonomic significance. IAWA Bulletin. 1991;12:143–175. [Google Scholar]
- Bello MA, Bruneau A, Forest F, Hawkins JA. Elusive relationships within order Fabales: Phylogenetic analyses using matK and rbcL sequence data. Systematic Botany. 2009;34:102–114. [Google Scholar]
- Bertioli DJ, Moretzsohn MC, Madsen LH, et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;23:10–45. doi: 10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso D, de Queiroz LP, Pennington RT, et al. Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. American Journal of Botany. 2012;99:1991–2013. doi: 10.3732/ajb.1200380. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Collier R, Fuchs B, Walter N, Lutke WK, Taylor CG. Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant Journal. 2005;43:449–457. doi: 10.1111/j.1365-313X.2005.02454.x. [DOI] [PubMed] [Google Scholar]
- Cota-Sánchez JH, Remarchuk K, Ubayasena K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Molecular Biology Reporter. 2006;24:161–167. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst AM, Jekat SB, Zielonka S, et al. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proceedings of the National Academy of Sciences of the USA. 2012;109:1980–1989. doi: 10.1073/pnas.1202999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. The phloem. In: Braun HJ, Carlquist S, Ozenda P, Roth I, editors. Encyclopedia of plant anatomy, Vol. 5. 2nd edn. Berlin: Borntraeger; 1969. 4–7. [Google Scholar]
- Esau K. Developmental features of the primary phloem in Phaseolus vulgaris L. Annals of Botany. 1978;42:1–13. [Google Scholar]
- Esau K, Cronshaw J. Tubular components in cells of healthy and tobacco mosaic virus-infected Nicotiana. Virology. 1967;33:26–35. doi: 10.1016/0042-6822(67)90090-6. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences of the USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi Y. Future directions of electron crystallography. Methods in Molecular Biology. 2013;955:551–568. doi: 10.1007/978-1-62703-176-9_30. [DOI] [PubMed] [Google Scholar]
- Furch ACU, Hafke JB, Schulz A, van Bel AJE. Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. Journal of Experimental Botany. 2007;58:2827–2838. doi: 10.1093/jxb/erm143. [DOI] [PubMed] [Google Scholar]
- Gould T, Harrington B, Hurst N, MenTaLgu Y. Inkscape. 2003. http://inkscape.org/ (3 March 2014)
- Groscurth S, Müller B, Schwan S, et al. Artificial forisomes are ideal models of forisome assembly and activity that allow the development of technical devices. Biomacromolecules. 2012;13:3076–3086. doi: 10.1021/bm3008499. [DOI] [PubMed] [Google Scholar]
- Hafke JB, Furch ACU, Fricker MD, van Bel AJE. Forisome dispersion in Vicia faba is triggered by Ca2+ hotspots created by concerted action of diverse Ca2+ channels in sieve elements. Plant Signaling & Behaviour. 2009;4:968–972. doi: 10.4161/psb.4.10.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaichi T, Sato T, Iwamoto T, Malavasi-Yamashiro J, Hoshino M, Mizuno N. A stable lead by modification of Sato's method. Journal of Electron Microscopy. 1986;35:304–306. [PubMed] [Google Scholar]
- Jäger M, Uhlig K, Clausen-Schaumann H, Duschl C. The structure and functionality of contractile forisome protein aggregates. Biomaterials. 2008;29:247–256. doi: 10.1016/j.biomaterials.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Jekat SB, Ernst AM, von Bohl A, et al. P-proteins in Arabidopsis are heteromeric structures involved in rapid sieve tube sealing. Frontiers in Plant Science. 2013;4:225. doi: 10.3389/fpls.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JAD. Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiology. 1974;53:96–103. doi: 10.1104/pp.53.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, van Bel AJE. Sieve tubes in action. Plant Cell. 1998;10:35–50. [Google Scholar]
- Knoblauch M, Peters WS, Ehlers K, van Bel AJE. Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell. 2001;13:1221–1230. doi: 10.1105/tpc.13.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, Noll GA, Müller T, et al. ATP-independent contractile proteins from plants. Nature Materials. 2003;2:600–603. doi: 10.1038/nmat960. Erratum in Nature Materials 4: 353. [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Stubenrauch M, van Bel AJE, Peters WS. Forisome performance in artificial sieve tubes. Plant, Cell & Environment. 2012;35:1419–27. doi: 10.1111/j.1365-3040.2012.02499.x. [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Systematic Biology. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lawton DM. Ultrastructural comparison of the tailed and tailless P-protein crystals respectively of runner bean (Phaseolus multiflorus) and garden pea (Pisum sativum) with tilting stage electron microscopy. Protoplasma. 1978a;97:1–11. [Google Scholar]
- Lawton DM. P-protein crystals do not disperse in uninjured sieve elements in roots of runner bean (Phaseolus multiflorus) fixed in glutaraldehyde. Annals of Botany. 1978b;42:353–361. [Google Scholar]
- Lewis G, Schrire B, Mackinder B, Lock M. Legumes of the world. Kew, UK: Royal Botanical Gardens; 2005. [Google Scholar]
- LPWG Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon. 2013;62:217–248. [Google Scholar]
- McDonald M, Box H, Bian W, Kendall A, Tycko R, Stubbs G. Fiber diffraction data indicate a hollow core for the Alzheimer's Aβ 3-fold symmetric fibril. Journal of Molecular Biology. 2012;423:454–61. doi: 10.1016/j.jmb.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon MM, Sanderson MJ. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionid legumes. Systematic Biology. 2006;55:818–836. doi: 10.1080/10635150600999150. [DOI] [PubMed] [Google Scholar]
- Mrazek A. Über geformte eiweiβartige Inhaltskörper bei den Leguminosen. Österreichische Botanische Zeitschrift. 1910;60:218–230. [Google Scholar]
- Müller B, Noll GA, Ernst AM, et al. Recombinant artificial forisomes provide ample quantities of smart biomaterials for use in technical devices. Applied Microbiology and Biotechnology. 2010;88:689–698. doi: 10.1007/s00253-010-2771-4. [DOI] [PubMed] [Google Scholar]
- Noll GA, Müller B, Ernst AM, et al. Characteristics of artificial forisomes from plants and yeast. Bioengineering Bugs. 2011;2:111–114. doi: 10.4161/bbug.2.2.14368. [DOI] [PubMed] [Google Scholar]
- Palevitz BA, Newcomb EH. The ultrastructure and development of tubular and crystalline P-protein in the sieve elements of certain papilionaceous legumes. Protoplasma. 1971;72:399–426. [Google Scholar]
- Pélissier HC, Peters WS, Collier R, van Bel AJE, Knoblauch M. GFP tagging of sieve element occlusion (SEO) proteins results in green fluorescent forisomes. Plant and Cell Physiology. 2008;49:1699–1710. doi: 10.1093/pcp/pcn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington RT, Lavin M, Ireland H, Klitgaard B, Preston J, Hu J-M. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Systematic Botany. 2001;26:537–556. [Google Scholar]
- Peters WS, Knoblauch M, Warmann SA, Schnetter R, Shen AQ, Pickard WF. Tailed forisomes of Canavalia gladiata: a new model to study Ca2+-driven protein contractility. Annals of Botany. 2007;100:101–109. doi: 10.1093/aob/mcm080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters WS, Knoblauch M, Warmann SA, Pickard WF, Shen AQ. Anisotropic contraction in forisomes: simple models won't fit. Cell Motility and the Cytoskeleton. 2008;65:368–378. doi: 10.1002/cm.20266. [DOI] [PubMed] [Google Scholar]
- Peters WS, Haffer D, Hanakam CB, van Bel AJE, Knoblauch M. Legume phylogeny and the evolution of a unique contractile apparatus that regulates phloem transport. American Journal of Botany. 2010;97:797–808. doi: 10.3732/ajb.0900328. [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Systematic Biology. 2005;54:441–454. doi: 10.1080/10635150590945359. [DOI] [PubMed] [Google Scholar]
- Pickard WF, Knoblauch M, Peters WS, Shen AQ. Prospective energy densities in the forisome, a new smart material. Material Science and Engineering: C. 2006;26:104–112. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Research. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana C, Cowley JM, Marhic C. Electron nanodiffraction and high–resolution electron microscopy studies of the structure and composition of physiological and pathological ferritin. Journal of Structural Biology. 2004;147:166–78. doi: 10.1016/j.jsb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Rüping B, Ernst AM, Jekat SB, et al. Molecular and phylogenetic characterization of the sieve element occlusion gene family in Fabaceae and non-Fabaceae plants. BMC Plant Biology. 2010;10:219. doi: 10.1186/1471-2229-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, et al. Genome structure of the legume, Lotus japonicus. DNA Research. 2008;15:227–39. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan S, Fritzsche M, Cismak A, Heilmann A, Spohn U. In vitro investigation of the geometric contraction behavior of chemo-mechanical P-protein aggregates (forisomes) Biophysical Chemistry. 2007;125:444–452. doi: 10.1016/j.bpc.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Schwan S, Menzel M, Fritzsche M, Heilmann A, Spohn U. Micromechanical measurements on P-protein aggregates (forisomes) from Vicia faba plants. Biophysical Chemistry. 2009;139:99–105. doi: 10.1016/j.bpc.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Stadler R, Wright K, Lauterbach C, et al. Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel postphloem domain in roots. Plant Journal. 2005;41:319–331. doi: 10.1111/j.1365-313X.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- Strasburger E. Über den Bau und die Verrichtungen der Leitungsbahnen in den Pflanzen. In: Fischer G, editor. Histologische Beiträge. Jena: Gustav Fischer; 1891. Book 3 193–195. [Google Scholar]
- Wergin WP, Newcomb EH. Formation and dispersal of crystalline P-protein in sieve elements of soybean (Glycine max L.) Protoplasma. 1970;71:365–388. [Google Scholar]
- Wergin WP, Palevitz BA, Newcomb EH. Structure and development of P-protein in phloem parenchyma and companion cells of legumes. Tissue and Cell. 1975;7:227–242. doi: 10.1016/0040-8166(75)90002-6. [DOI] [PubMed] [Google Scholar]
- Will T, van Bel AJE. Physical and chemical interactions between aphids and plants. Journal of Experimental Botany. 2006;57:729–737. doi: 10.1093/jxb/erj089. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. American Journal of Botany. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- Zee S-Y. Fine structure of the differentiating sieve elements of Vicia faba. Australian Journal of Botany. 1969;17:441–456. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.