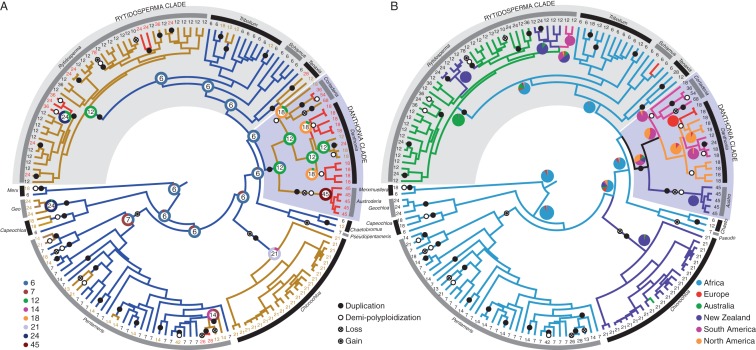
Fig. 2.
Maximum clade credibility (MCC) tree topology of 154 species of Danthonioideae for which ploidy data are available, based on 14 425 aligned DNA base pairs. The branches are scaled to age. Whole-genome duplications are indicated as filled circles (either one or more duplications), demi-polyploidizations by open circles, losses of one or more chromosomes by circles with a plus, and gains of one or more chromosome by a circle with a central dot; species names have been replaced by the haploid chromosome number. (A) The most likely haploid chromosome numbers are indicated at the critical nodes, inside a doughnut indicating the posterior probabilities of all haploid numbers at that node. Branch colours reflect the ploidy level: the ancestral haploid number is six; nodes that retain this ploidy level are coloured blue. A first increase in ploidy is coloured brown, and a second increase (either duplication or demi-polyploidization) red. (B) Branch colours indicate the geographical regions as inferred from ML, and the probability distribution (as determined by stochastic mapping) at critical nodes is given as pie-charts. Abbreviations of generic names: Austro, Austroderia; Chaeto, Chaetobromus; Geo, Geochloa; Merx, Merxmuellera; Pseudo, Pseudopentameris.