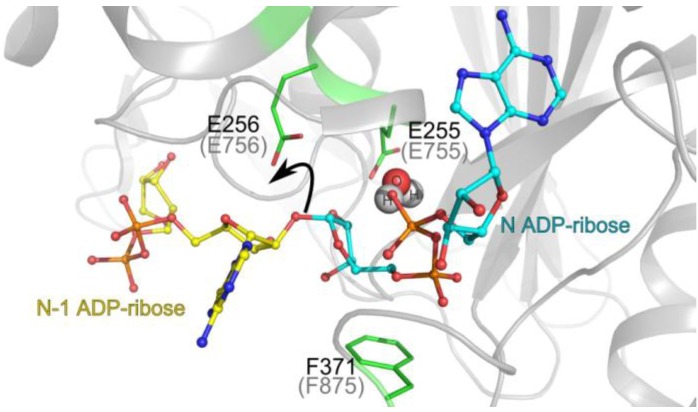
Figure 3.
PARG catalytic mechanism. The positions of the reaction product N ADP-ribose (teal backbone; observed in the crystal structure) and a model of N-1 ADP-ribose (yellow) are represented in the Tetrahymena thermophila PARG active site. The key O-glycosydic ribose-ribose bond is positioned in direct hydrogen bonding contact with the catalytic glutamate 256 (Glu756 in human PARG; grey brackets). The 2’-OH leaving group of n-1 ADP-ribose’ is protonated (black arrow) by the catalytic glutamate. A subsequently formed oxocarbenium intermediate is stabilised by the close proximity of the terminal diphosphate group, which is restrained by the conserved phenylalanine 371 (Phe875 in human PARG). A water molecule (shown in spheres) is ideally positioned to attack this oxocarbenium intermediate. This leads to release of ADP-ribose.