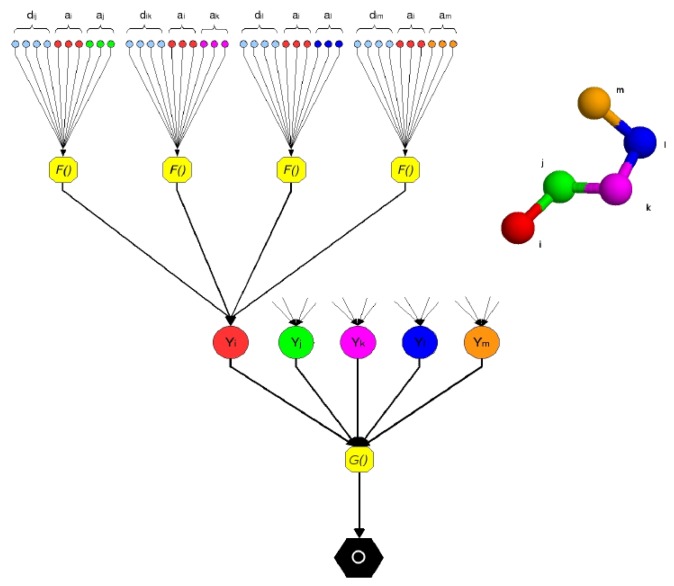
Figure 1.
A pentapeptide structure is processed by the Neural Network Pairwise Interaction Fields (NNPIF). The structure is represented as a directed acyclic graph (DAG) by considering as leaf nodes the interactions between couples of atoms. Each interaction is modeled by the function, F(), via an NN, NF. The inputs encode characteristics of each of the two interacting atoms (ax, ay) in the pair and their interaction (dxy). The outputs are combined in hidden vectors Y. The contribution of each interaction to the quality of the structure is modeled by the function, G(), via an NN, NG, which produces the global output, O.