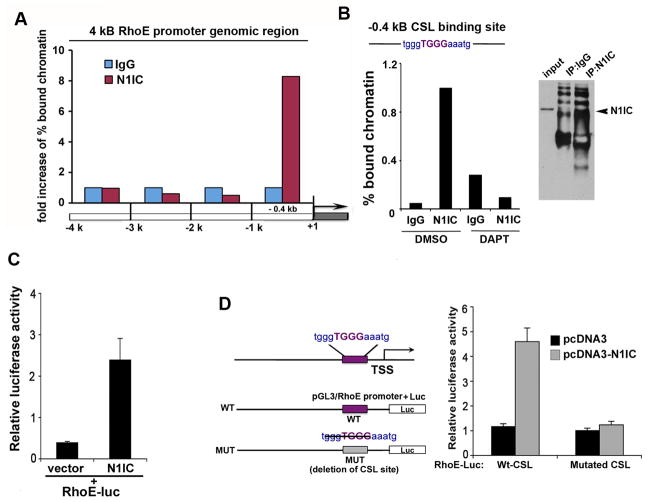
Figure 3. Identification of RhoE as a transcriptional target of Notch1.
A, Endogenous Notch1 is enriched at −1kb genomic region from the transcription start site (TSS) of human RhoE. Primary HKC were forced to differentiate in suspension and processed for scanning ChIP of the −5kb genomic region upstream of the human RhoE TSS, as described in the Supplementary Information, using an antibody against the activated form of Notch1 or purified rabbit IgGs. B, Differentiated primary HKC, teraetd with DMSO control or DAPT were processed for ChIP as in (A) followed by real time PCR analysis (upper panel) or conventional PCR (lower pane) using specific primers (described in Supplementary Information) to amplify the −0.4kb region of the human RhoE promoter containing the CSL binding site. Efficiency of the immunoprecipitation is shown in the right panel: Western blot analysis of immunoprecipitates performed either with non-specific IgG or with the antibody against the activated form of Notch1. C, SCC028 cells were transfected with a N1IC construct or empty vector together with the RhoE reporter (RhoE-luc) and phRL-TK Renilla reporter. D, The promoter regions were analyzed using the rVista software for identification of transcription factor binding sites. The CSL binding site at position −0.4 kb was deleted, and SCC028 keratinocyte cells were transfected with the mutated or with the wild type luciferase reporter constructs together with the NICD construct and phRL-TK Renilla reporter. Error bars represent Standard Deviations, calculated based on three independent measurements.