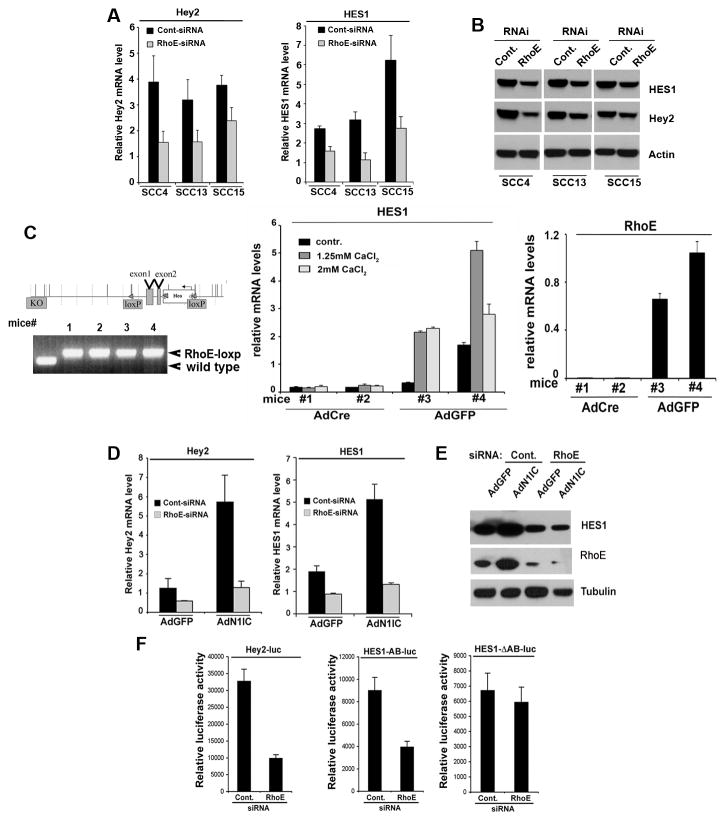
Figure 4. Depletion of RhoE inhibits downstream Notch signaling.
A, SCC4, SCC13 and SCC15 cancer cell lines were transfected with control and RhoE siRNA and the levels of the Notch target genes Hey2 and HES1 were analyzed by real time RT-PCR analysis using 36B4 as an internal control. B, Protein levels of Hey2, HES1 and RhoE in the same cells were analyzed by Western blot using actin levels as an internal loading control. C, Inducible keratinocyte-specific deletion of the RhoE gene and resulting effects on Notch signaling and keratinocyte differentiation. Left upper panel: Schematic representation of the murine RhoE gene and of the expected recombination products leading to its inducible tissue-specific deletion. After induction of Cre recombinase activity (the loxP sites are indicated with red arrows), the genomic portion harboring the first two exons are deleted. Left lower panel: PCR analysis of genomic DNA of offspring from the RhoE loxp/loxp homozygous mice: the primers amplify a larger fragment including the loxp site and wild type genomic DNA is presented as a control. Middle panel: Real-time RT-PCR analysis of mRNA isolated from primary keratinocytes derived from mutant RhoE loxp/loxp mice and infected with an adenovirus expressing the Cre recombinase or an Ad-GFP control. At 2 days after infection levels of Hes1 are decreased in cells with Cre activity upon calcium mediated induction of differentiation. Depletion of RhoE mRNA levels shown on the right. D, Primary HKC under growing adherent conditions were transfected with control (scrambled siRNA) or RhoE siRNA, and 48 hours later were infected with adenoviruses expressing either GFP (AdGFP) or the activated form of Notch1 (AdN1IC). 24 hours later expression of Notch1 target genes, Hey2 and HES1 was analyzed by real time RT-PCR. E, RhoE and HES1 expression in the same cells was analyzed by Western blot. F, SCC028 cells transfected with control or RhoE siRNA were transfected with luciferase reporters for Hey2 (Hey2-luc, 0.5mg), wild type HES1 containing both CSL binding sites in the promoter region (HES1-AB-luc, 0.5mg) or HES1-ΔAB-luc with mutated CSL binding sites together with a Renilla minimal reporter (0.05 mg) for internal normalization. 24 hrs after transfection, luciferase activity was measured and normalized with Renilla activity. Error bars represent Standard Deviations, calculated based on three independent measurements.