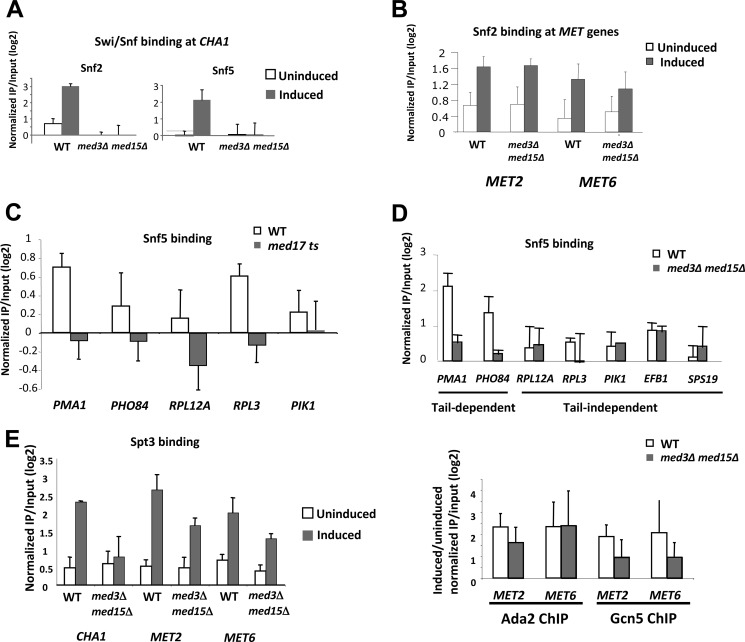
FIGURE 2.
Dependence on Mediator of Swi/Snf and SAGA association with active gene promoters. A, Snf2 and Snf5 association with the CHA1 promoter in wild type (WT; BY4742 for Snf5 and Research Genetics Snf2-TAP strain for Snf2) and gal11/med15Δ med3Δ (LS10 for Snf5 and RMY525 for Snf2) yeast grown in CSM without (uninduced) or with 1 mg/ml of serine (induced) was measured by ChIP. B, Snf2 association with the MET2 and MET6 promoters in WT (Snf2-TAP strain, Research Genetics) and gal11/med15Δ med3Δ (RMY525) yeast grown in YPD without (uninduced) or with (induced) 0.5 mm CdCl2. C, Snf5 association with the indicated promoters was measured in WT (RMY521) and srb4/med17 ts (RMY522) yeast grown in YPD after 1 h at 37 °C. D, Snf5 association with promoters whose transcription is dependent or independent of the Mediator tail module, as indicated, was measured in WT (Snf2-TAP strain, Research Genetics) and gal11/med15Δ med3Δ (RMY525) yeast grown in YPD. E, effect of Mediator tail mutation on SAGA recruitment. Left panel: Spt3 association with the CHA1 promoter was measured by ChIP in wild type (Snf2-TAP strain, Research Genetics) and gal11/med15Δ med3Δ yeast (RMY525) grown in CSM without (uninduced) or with (induced) 1 mg/ml of serine, and association with the MET2 and MET6 promoters was measured in wild type (RMY521) and gal11/med15Δ med3Δ yeast (RMY415) grown in YPD without (uninduced) or with (induced) 0.5 mm CdCl2. Right panel, Ada2 and Gcn5 association at the uninduced and induced MET2 and MET6 promoters were measured by ChIP, and the log2 ratios of the association at the induced/uninduced promoters in wild type and gal11/med15Δ med3Δ yeast are shown, as indicated. Relative association in all cases was determined by quantitative PCR analysis of input and immunoprecipitated (IP) samples, and normalized to a non-transcribed region of ChrV (25). Error bars represent S.D. (n = 3–4).