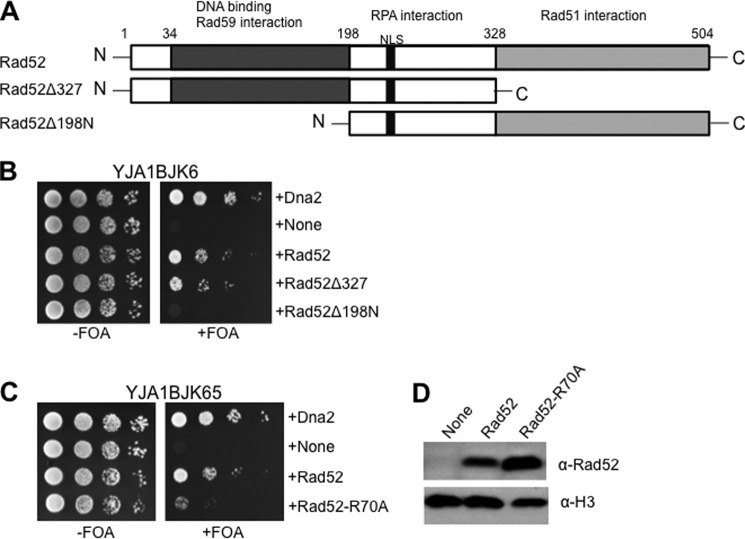
FIGURE 6.
The Rad52-dependent suppression of dna2-K1080E depends on the DNA annealing activity of Rad52. A, schematic representation of Rad52. Domains required for DNA binding and interactions with Rad59, RPA, and Rad51 are as shown. The two truncated fragments, Rad52Δ327 and Rad52Δ198N, are as indicated. NLS, nuclear localization signal. B, the YJA1BJK56 strain was transformed with each of the five pRS325-ADH1 plasmids expressing Dna2, empty vector (None), Rad52, Rad52Δ327, and Rad52Δ198N. The resulting transformants were grown and spotted as described in the legend for Fig. 1A. C, the pRS325-ADH1 plasmids expressing the proteins indicated at the right of the figure were transformed into YJA1BJK65. Transformants were grown and spotted as described in the legend for Fig. 1A. D, expression levels of Rad52 and Rad52-R70A proteins were examined as described in legend for Fig. 1B. Histone H3 was used as the loading control. α-H3, antibodies against histone H3.